Original Article - DOI:10.33594/000000755
Accepted 24 December 2024 - Published online 21 January 2025
MOTS-c Impact on Muscle Cell Differentiation and Metabolism Across Fiber Types
Keywords
Abstract
Background/Aims:
MOTS-c belongs to a group of mitochondrial peptides involved in metabolic processes in the body. This peptide has garnered increasing attention since its discovery in 2015 because of its potential to ameliorate metabolic parameters in animals with diabetes or insulin resistance. MOTS-c is involved in muscle metabolism; however, little is known about its role in fiber differentiation.Materials:
We conducted a study to explore the effect of MOTS-c on cellular processes using the C2C12 and L6 cell lines, representing different metabolic types of muscle fibers. The research methods were real-time PCR, Western blot, and lipid accumulation measurement.Results:
Notably, our investigations revealed that MOTS-c increased the survival of C2C12 cells at doses of 10 and 100 nM (p<0.01) and stimulated the phosphorylation of extracellular signal-regulated kinase within 5 min of incubation (p<0.05). Remarkably, these effects were not observed in L6 cells; however, both cell lines showed a reduced rate of proliferation. Furthermore, MOTS-c promotes the differentiation of C2C12 cells by increasing the expression of muscle regulatory factors, but it does not produce such an effect in L6 cells. Additionally, cells were treated with physiological concentrations of free fatty acids and MOTS-c, unveiling an augmentation in lipid accumulation observed in L6 cells and a decrease in lipid accumulation in C2C12 cells.Conclusion:
In conclusion, our findings have suggested a diverse response to MOTS-c depending on the type of muscle fibers, particularly in the domains of survival, cell differentiation, and lipid accumulation.Introduction
Cell differentiation demands a massive amount of energy investment—the cell undergoes shape modifications and a complete reconstruction of its protein composition. A cell activated for differentiation loses its potential for further differentiation, although this potential might be preserved in the initial stage of differentiation. In the muscle tissue, satellite cells play a role in generating new muscle fibers. Positioned at the periphery of the fibers, they become activated in response to factors such as tissue damage. In the postnatal period, the satellite cells constitute approximately 30% of all cells in the muscle tissue. In adulthood, these cells account for only 5% [1]. A definitive marker of satellite cells is the expression of the paired box 7 (PAX7) transcription factor, which becomes inactive upon initiating the cell differentiation process. During differentiation, protein factors, known as muscle regulatory factors (MRFs), precisely guide the cell from the myoblast to the myotube [2]. Upon activation, myogenic factor 5 (myf5) protein expression was increased. During early differentiation, the myoblast determination protein 1 (myoD) factor participates in the early stage of differentiation, thus curbing cell proliferation by repressing Myf5 expression and activating myogenin expression [3]. Subsequently, muscle regulatory factor 4 (MRF4) collaborates with myogenin to shape multinuclear tubes [3]. These intricate processes, as previously mentioned, demand substantial energy expenditure. Notably, differentiation significantly increases mitochondrial enzymatic activity [4].
Mitochondria transcend their role as energy sources and producers of diverse peptides termed mitochondrial-derived peptides (MDPs) [5]. The mitochondrial open reading frame of the 12S rRNA-c (MOTS-c), an MDP, is a relatively understudied peptide comprising 16 amino acids [6], and it was discovered in 2015. MOTS-c is a vital indicator of cellular metabolic state [7]. This peptide has been demonstrated to promote cellular glucose uptake and enhance fatty acid oxidation in the adipocytes [8, 9]. However, the interplay between MOTS-c and other tissues remains an actively developing area of research. While MOTS-c is a peptide of great importance for cellular metabolism, its function in muscle cell differentiation remains inadequately understood. MOTS-c enhances muscle performance and diminishes myostatin expression [10], suggesting a growth-promoting effect on the skeletal muscle tissue. Therefore, this study aimed to investigate the influence of MOTS-c on the physiological features of muscle fibers in an in vitro model using two cell lines, namely, C2C12 and L6.
Materials and Methods
Cell culture and cell differentiation
The murine C2C12 and rat L6 cell lines were purchased from the European Collection of Authenticated Cell Cultures. Cells were cultured in a growth medium Dulbecco’s Modified Eagle Medium (DMEM) containing 4.5 g/L glucose and L-glutamine, supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The cells were maintained at 37°C in a humidified 5% CO2 incubator. When the cells reached 90% confluence, the growth medium was replaced with a differentiation medium: DMEM supplemented with 2% horse serum [11]. The medium was replenished with fresh medium at 2-day intervals throughout the experiments.
Cell viability
We performed the 3-[4, 5-dimethylthiazol-2-yl]-2, 5 di-phenyl tetrazolium bromide (MTT) assay to evaluate cell viability. This assay is based on living cells converting MTT into formazan crystals. Cells were seeded into 96-well plates containing growth medium and cultured for 24 h. The growth medium was then replaced with an experimental medium supplemented with 0.2% bovine serum albumin (BSA), and the cells were exposed to various concentrations of MOTS-c (1, 10, and 100 nM). After 24 h, MTT solution was added and incubated for 1 h. Subsequently, the medium was removed, and the resulting crystals were dissolved with 100 µl of dimethyl sulfoxide. The absorbance was detected using a Synergy 2 microplate reader (BioTek Instruments, Winooski, VT, USA).
RNA isolation and qRT-PCR
Total RNA was isolated using the EXTRAzol reagent (Gdansk, Poland) according to the manufacturer’s instructions. Briefly, 1 µg of total RNA was used to obtain cDNA. Reverse transcription of the cDNA was performed using a High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Grand Island, NY, USA). Real-time polymerase chain reaction (PCR) was performed using QuantStudio™ 12K Flex™ with 5× HOT FIREPol® Eva-Green® qRT-PCR Mix Plus (ROX) and gene-specific primers. The specificity of the PCR assay products was ascertained by determining the melting points (0.1°C/s transition rate). Relative gene expression was quantified using the 2−ΔΔCt method, with GAPDH as the reference gene. Primer sequences used in the qRT-PCR assay are listed in Table 1.

Table 1: Primer sequences used in the qRT-PCR assay
Protein isolation
Total proteins were obtained using a radioimmunoprecipitation assay buffer (RIPA). Protein concentration was measured using a BCA protein concentration assay kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
ERK1/2 phosphorylation
For western blotting analysis, cells were seeded into 6-well plates containing a growth medium for 24 h. Following removing the medium, the cells were washed with phosphate-buffered saline (PBS). The medium was replaced with an experimental medium supplemented with 0.2% for 3 h. The medium was again replaced with a medium supplemented with 100 nM MOTS-c, wherein cells were treated for 5, 10, 15, 30, and 45 min. The cells were then lysed immediately in cold radioimmunoprecipitation assay buffer, after which the lysate was transferred to tubes and centrifuged at 13000×g for 10 min. The supernatant was collected for protein isolation, and the protein concentration was quantified using the bicinchoninic acid method. The resulting protein sample was used for the subsequent western blot analysis at −80°C.
Western blot assay
The protein sample (30 µg) was loaded and resolved on a 12% Tris-HCl SDS-PAGE gel, and the separated proteins were transferred onto a polyvinylidene difluoride membrane. The blotted membranes were incubated overnight at 4°C with primary antibodies. After incubation, the membranes were washed thrice with Tris-buffered saline buffer, and the membranes were incubated with secondary antibodies for 1 h. Signals were visualized using the ChemiDoc system (Bio-Rad, USA).
Lipid accumulation and ORO staining
Oil red O (ORO) staining was performed on differentiated myotubes. Cells were seeded in 96-well plates containing a growth medium. After reaching 90% confluence, the cells were differentiated using a differentiation medium for 6 days, with medium replacement occurring every second day.
Subsequently, myotubes were treated with PBS, and the medium was supplemented with 100 nM MOTS-c alongside 0.2% BSA and free fatty acids, including oleic, stearic, and palmitic acid at concentrations of 200 µM. Fatty acids were conjugated with BSA overnight before the experiment [12]. After 24 h, cells were fixed with 10% paraformaldehyde, followed by ORO staining.
The ORO working solution was prepared by mixing the ORO solution and distilled water in a 6:4 ratio. Following a 20-minute incubation, the solution was filtered using a 0.2-µm syringe filter. During this time, the cells were rinsed with distilled water, followed by a 60% isopropanol solution, and subsequently dried. The ORO working solution was then applied for 10 min. The cells were immediately rinsed thrice with distilled water. The cells were wholly dried and dissolved in 100% isopropanol for quantification. Absorbance was read using a Synergy 2 Multi-Mode Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA).
Acetate Uptake
C2C12 or L6 cells were seeded and differentiated in 12-well plates as described above. Next, cells were incubated in a Krebs-Ringer phosphate buffer (KRB) (25 mM HEPES [pH 7.4], 118 mM NaCl, 4.8 mM KCl, 1.3 mM CaCl2, 1.2 mM KH2PO4, 1.3 mM MgSO4, 5 mM NaHCO3, 0.2% bovine serum albumin, and 5.5 mM glucose). After the preincubation, cells were incubated for 30 min in Krebs-Ringer buffer in the presence or absence of MOTS-c (100 nmol/L). Next, KRB buffer with C14-acetate sodium (18 kBq/μmol) was added, and cells were incubated for 20 min. Then, ice-cold PBS was added. The cells were lysed using 0.1% SDS. Lysates were transferred into scintillation liquid, and β-radiation was measured.
Lipogenesis
Lipogenesis was measured by the evaluation of C14-acetate sodium incorporation into lipids. C2C12 or L6 cells seeded and differentiated in 12-well plates as described above. After differentiation, cells were treated with or without MOTS-c (100 nmol/L) in KRB buffer containing 0.1% BSA. KRB was supplied with 10 µM sodium acetate and C14-acetate per well to stimulate lipogenesis. Myotubes were incubated for 16 h at 37°C in lipogenesis medium. After incubation, Dole’s extraction mixture was added to stop the reaction. Subsequently, H2O and heptane were added, and the upper phase containing the lipid fraction was transferred to scintillation liquid for b-counting.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA, USA). One-way analysis of variance was applied for calculations, supplemented by a Dunnett post hoc test (in comparison with the control group) or Tukey post hoc test (for intergroup comparisons), alongside unpaired Student’s t-test (two-tailed distribution) for *p<0.05 and **p<0.01. The results are presented as the arithmetic mean±SEM.
The murine C2C12 cell line and the rat L6 cell line were purchased from the European Collection of Authenticated Cell Cultures. Cells were cultured in a growth medium (Dulbecco’s Modified Eagle Medium (DMEM) containing 4.5 g/L glucose and L-glutamine), supplemented with 10% fetal bo-vine serum and 1% penicillin–streptomycin. The cells were maintained at 37°C in a humidified 5% CO2 incubator. When the cells reached 90% confluence, the growth medium was replaced with a differentiation medium: DMEM supplemented with 2% horse serum [11]. Throughout the experiments, the medium was replenished with fresh medium at 2-day intervals.
Results
Effect on viability and ERK phosphorylation
We investigated the effect of the MOTS-c peptide on the survival of the tested cell lines. MOTS-c could increase survival in C2C12 cells (10 and 100 nM; both p<0.01; Fig. 1A), yet no discernible effect was noted on survival in L6 cells (Fig. 1B). We then assessed cell proliferation based on proliferating cell nuclear antigen (PCNA) protein expression. We observed that MOTS-c exerted an inhibitory effect on the proliferation of both C2C12 cells (100 nM, p<0.05; Fig. 1C) and L6 cells (10 nM, p<0.01; Fig. 1D). In the subsequent investigation into the effect of MOTS-c on the phosphorylation of extracellular signal-regulated kinases (ERK), we found an increase in ERK kinase phosphorylation in the C2C12 line after 5 and 10 min (p<0.05; Fig. 1E). Conversely, no alterations in ERK kinase phosphorylation were identified in L6 cells.
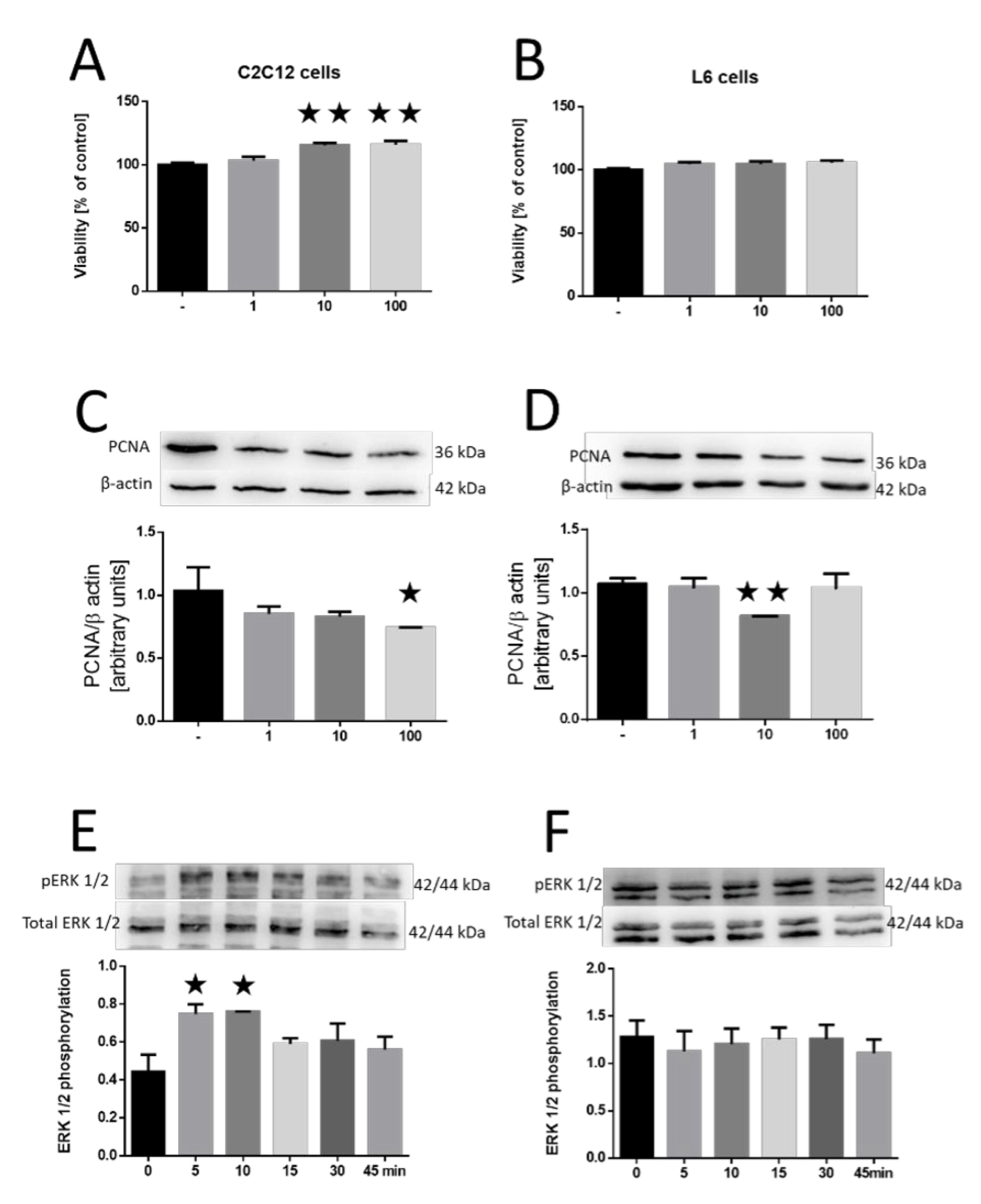
Fig. 1: Effect of MOTS-c on the survival of C2C12 (A) and L6 (B) cell lines. Changes in PCNA expression after incubation with MOTS-c in C2C12 (C) and L6 (D) cell lines. The process of ERK1/2 kinase phosphorylation after the addition of MOTS-c in C2C12 (E) and L6 (F) cell lines. The results are shown as a percentage of the control group (set to 100%), as mean±standard error of the mean (SEM). Statistically significant differences are represented as *p<0.05 and **p<0.01 versus the corresponding control group using a one-way analysis of variance followed by Dunnett’s post hoc test.
Effect on differentiation in C2C12 cells
To assess the effect of MOTS-c on cell differentiation, we examined the expression of genes related to MRFs. Our findings indicated an upregulated expression of myogenin after 48 h (10 nM, p<0.05; Fig. 2A) and myoD (10 nM, p<0.05; Fig. 2B). At the protein level, we observed an increase in the expression of myogenin (100 nM, p<0.05; Fig. 2C) and myoD (1 and 10 nM, both p<0.05; 100 nM, p<0.01; Fig. 2D). Further exploration into the expression of myosin chain genes yielded no significant effect of MOTS-c at any tested dose after 48 h of incubation (Fig. 2E and F).
The same parameters were re-evaluated with the tested peptide after a 6-day incubation period. Our findings revealed elevated myogenin gene expression (10 nM, p<0.01; Fig. 3A) and an increase in myoD (10 nM, p<0.05; Fig. 3B). The increase in these factors mirrored at the protein level—myogenin expression was upregulated at all tested doses of MOTS-c (1 and 100 nM, p<0.01; 10 nM, p<0.05; Fig. 3C), while myoD expression exhibited an increase at 10 and 100 nM doses (10 nM, p<0.05; 100 nM, p<0.01; Fig. 3D). After the 6 days, we observed amplified myosin gene expression in both myosin heavy chain 3 (Myh3) (Fig. 3E) and myosin heavy chain 4 (Mhc4) at the 10-nM dose (10 nM, p<0.05; Fig. 3F).
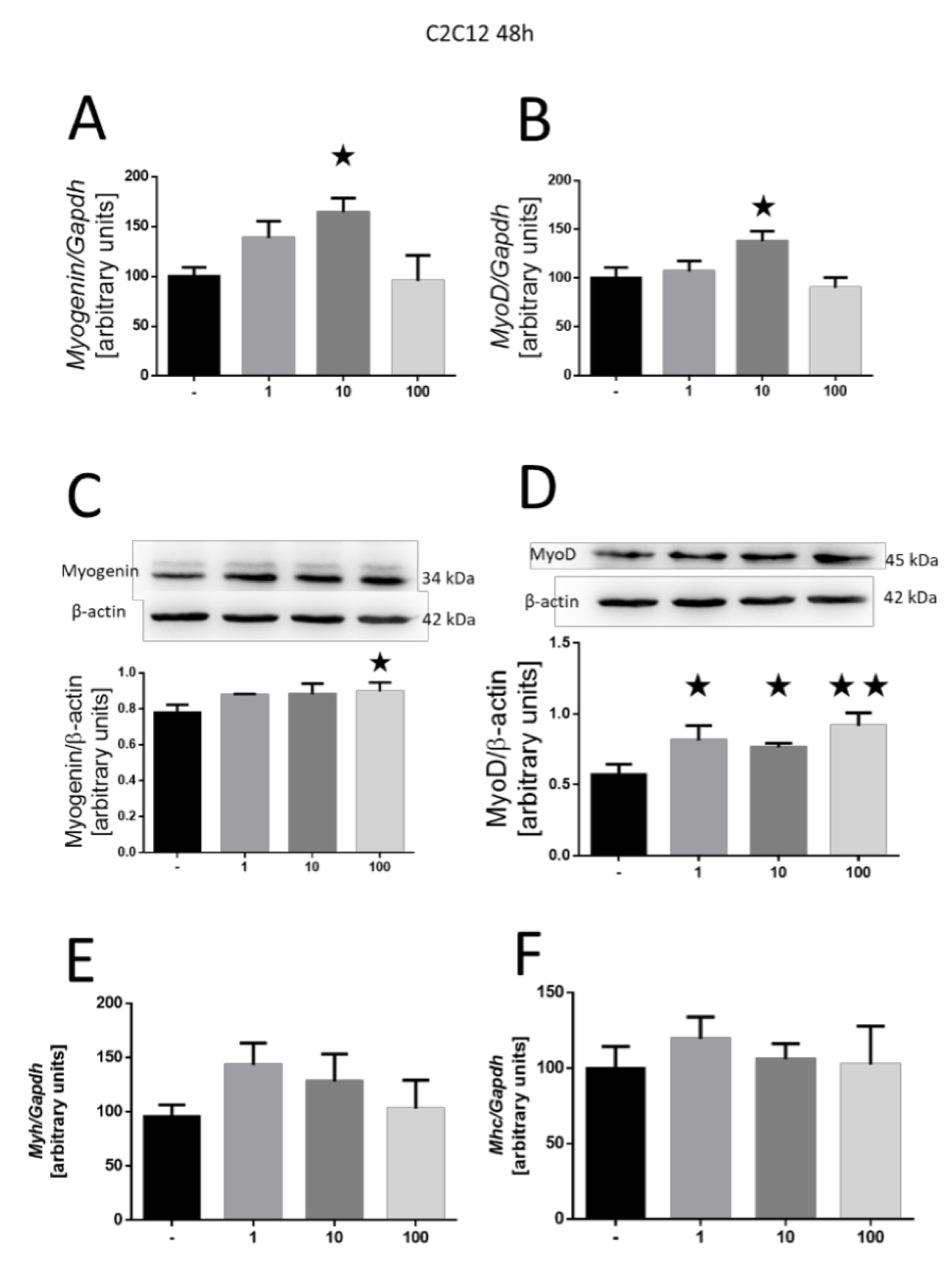
Fig. 2: Effect of MOTS-c on the differentiation of C2C12 cells after incubation for 2 days. Expression-related changes in myogenin (A) and myoD (B), and gene and protein (C, D), respectively. Expression of Myh3 (E) and Mhc4 (F) myosin chains. The results are shown as a percentage of the control group (set to 100%), as mean±SEM. Statistically significant differences are represented as *p<0.05 and **p<0.01 versus the corresponding control group using a one-way analysis of variance followed by Dunnett’s post hoc test.
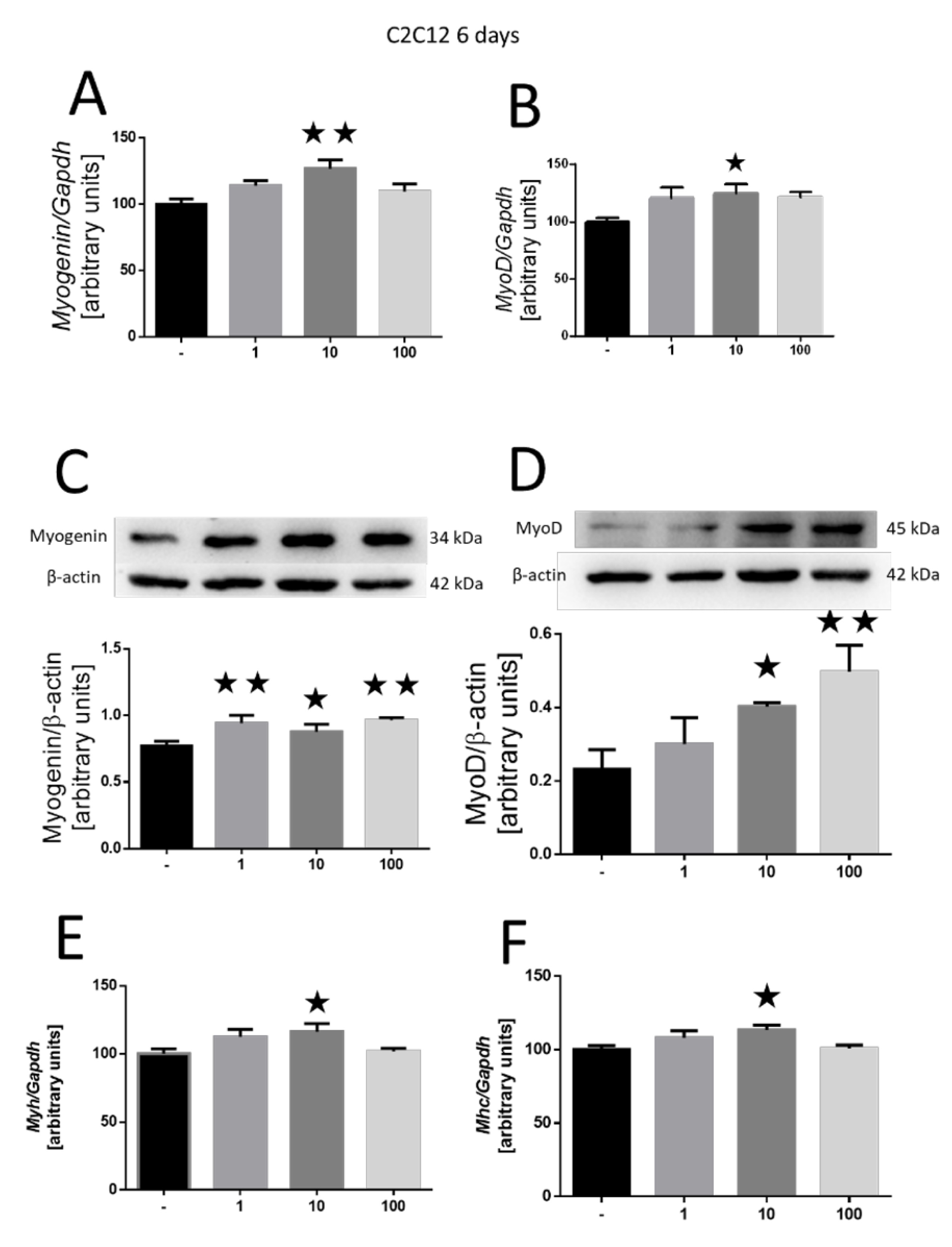
Fig. 3: Effect of MOTS-c on the differentiation of C2C12 cells after 6 days of incubation. Expression-related changes in myogenin (A) and myoD (B), and gene and protein (C, D), respectively. Changes in the expression of Myh3 (E) and Mhc4 (F) myosin chains. The results are shown as the percentage of the control group (set to 100%), as mean±SEM. Statistically significant differences are represented as *p<0.05 and **p<0.01 versus the corresponding control group using a one-way analysis of variance followed by Dunnett’s post hoc test.
Effect on differentiation in L6 cells
The investigation into cell differentiation extended to the L6 cell line. For the 48-h incubation period, no alteration in myogenin expression was detected (Fig. 4A). However, an increase in myoD expression was observed (100 nM, p<0.05; Fig. 4B). There are no changes in myogenin protein expression (Fig. 4C) and myoD (Fig. 4D). Expression analysis of myosin chains indicated an increase in Mhc4 expression (10 nM, p<0.05; Fig. 4F), while Myh3 gene expression remained unchanged (Fig. 4E).
The examination continued after 6 days of incubation, yet no effect of MOTS-c on L6 cell lineage differentiation was demonstrated. No changes in myogenin gene expression (Fig. 5A) and myoD (Fig. 5B) were observed. Additionally, no changes were observed in myogenin protein expression (Fig. 5C) or myoD (Fig. 5D). Similarly, the expression of Myh3 (Fig. 5E) and Mhc4 (Fig. 5F) myosin chain genes remained unaffected.
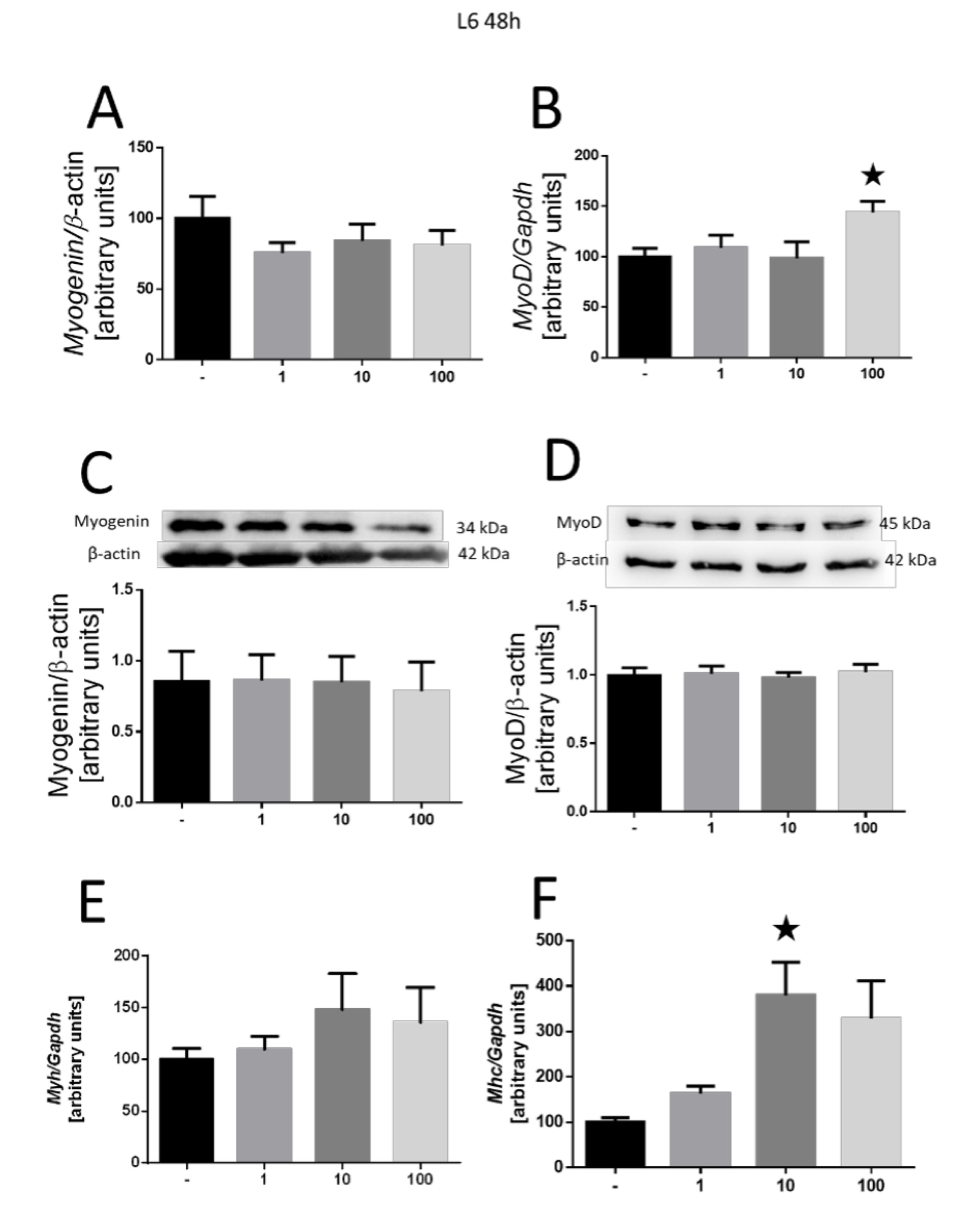
Fig. 4: Effect of MOTS-c on the differentiation of L6 cells after 2 days of incubation. Changes in myogenin (A) and myoD (B), and gene expression and protein (C, D), respectively. Expression-related changes in the Myh3 (E) and Mhc4 (F) myosin chains. The results are shown as a percentage of the control group (set to 100%), as mean±SEM. Statistically significant differences are presented as *p<0.05 versus the corresponding control group using a one-way analysis of variance followed by Dunnett’s post hoc test.
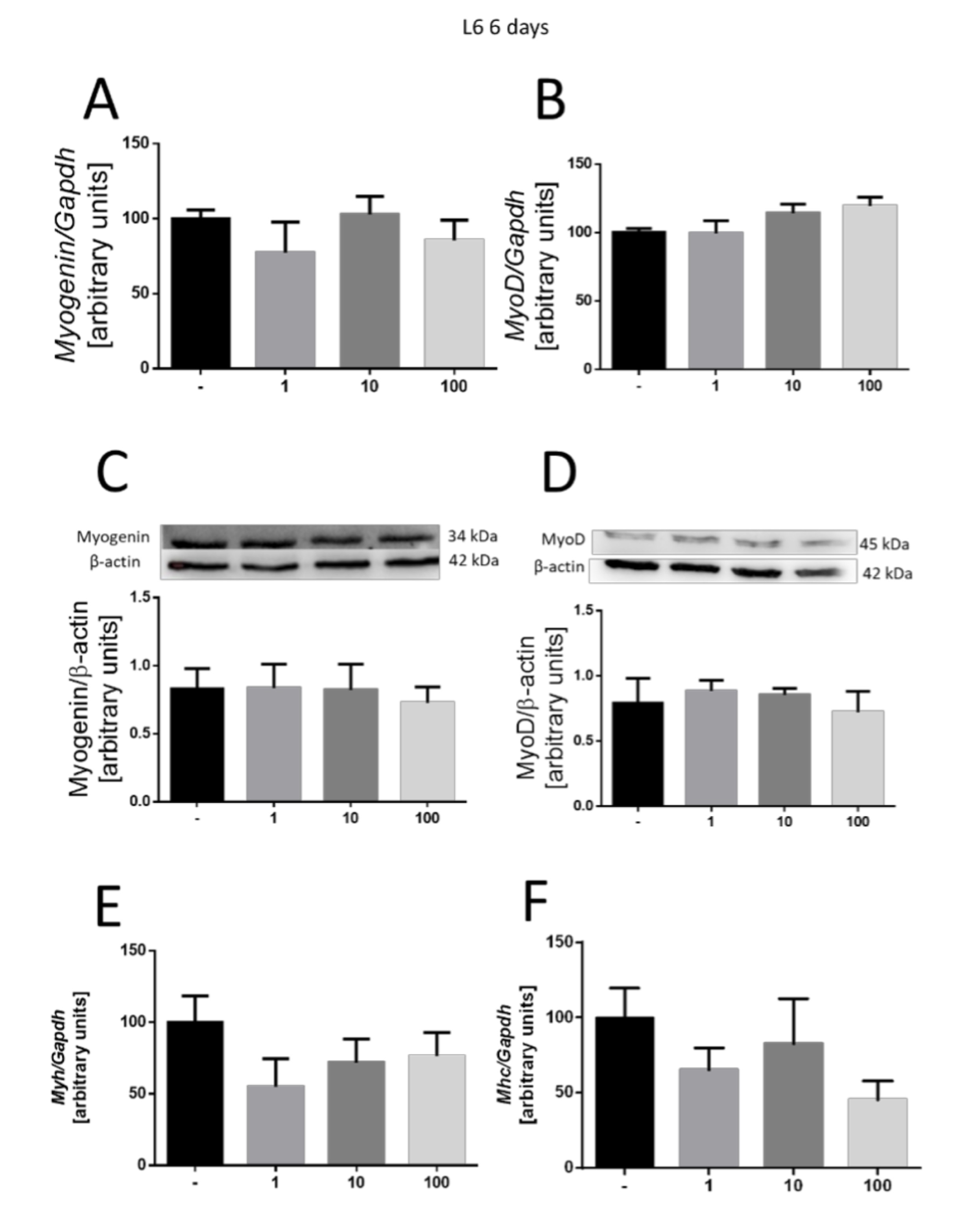
Fig. 5: Effect of MOTS-c on the differentiation of L6 cells after 6 days of incubation. Expression-related changes in myogenin (A) and myoD (B), and gene and protein (C, D), respectively. Changes in the expression of Mhc4 (E) and Myh3 (F) myosin chains. The results are shown as a percentage of the control group (set to 100%), as mean±SEM.
Effect of MOTS-c on lipid accumulation in tested cell lines
Subsequently, we elucidated the effect of MOTS-c influence on lipid accumulation in fully differentiated C2C12 and L6 myotubes. After 24 h of incubation with fatty acids and MOTS-c, C2C12 cells exhibited diminished stearic acid accumulation (p<0.001), oleic acid (p<0.01), and fatty acid mixture (p<0.01). Notably, MOTS-c did not affect palmitic acid accumulation (Fig. 6A). Contrastingly, the L6 cell line demonstrated more accumulation of stearic acid (p<0.05), oleic acid (p<0.05), and palmitic acid (p<0.01). The acid mix showed no significant changes (Fig. 6B).
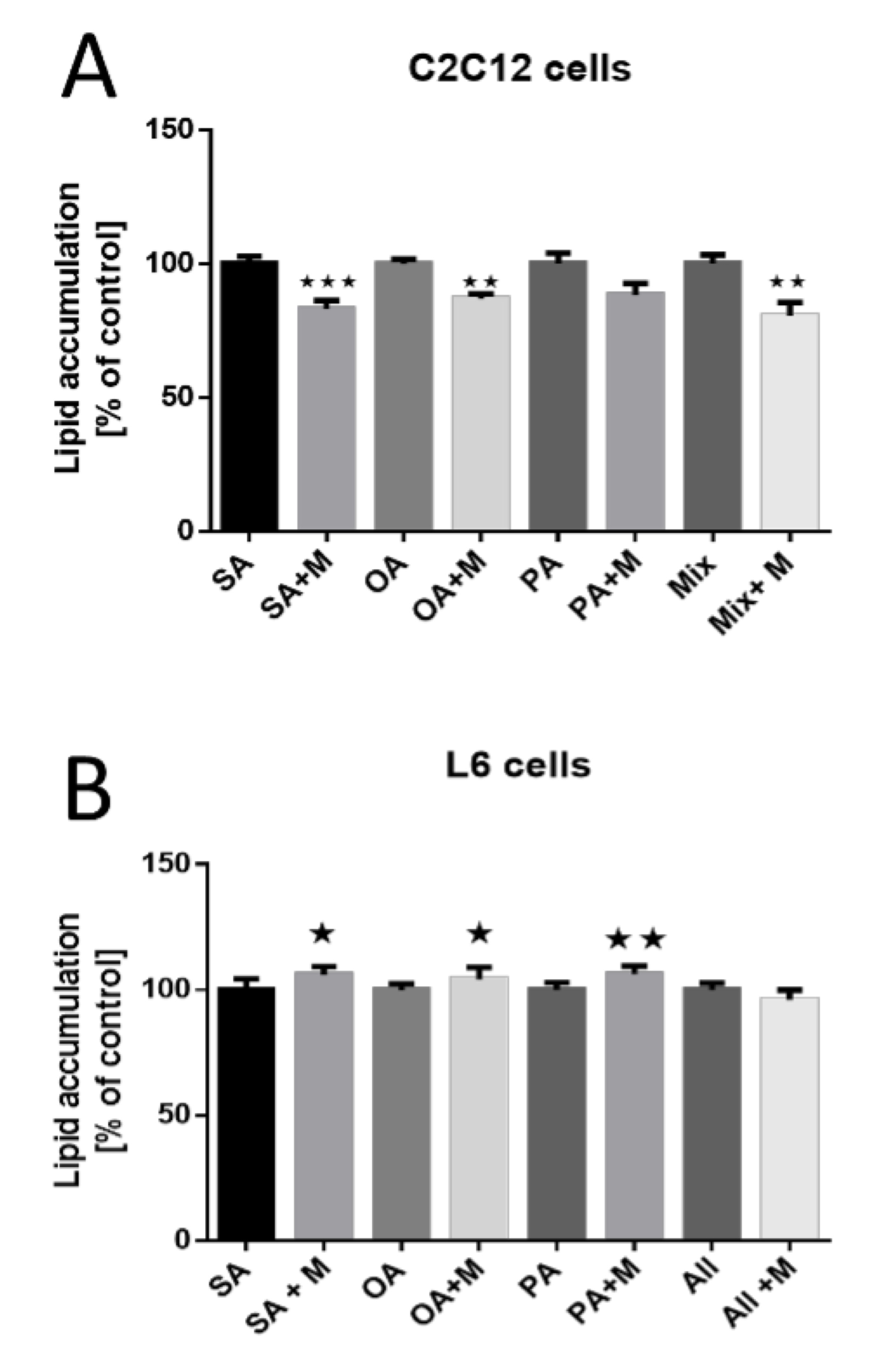
Fig. 6: Effect of MOTS-c on lipid accumulation in C2C12 (A) and L6 (B) muscle cells. The results are shown as a percentage of the control group (set to 100%), as mean±SEM. Statistically significant differences are represented as *p<0.05, **p<0.01, ***p<0.001 versus the corresponding control group using unpaired Student’s t-test (two-tailed distribution).
Effects of MOTS-c on lipogenesis and sodium acetate uptake
The impact of MOTS-c on lipogenesis in both C2C12 and L6 cell lines showed no significant changes (Figures 7A and 7B). However, when examining sodium acetate uptake, a significant reduction was observed in C2C12 cells (*p<0, 05; Fig. 7C), while L6 cells exhibited no notable changes in uptake (Fig. 7D). These findings underscore the contrasting metabolic responses of the two cell lines to MOTS-c treatment.
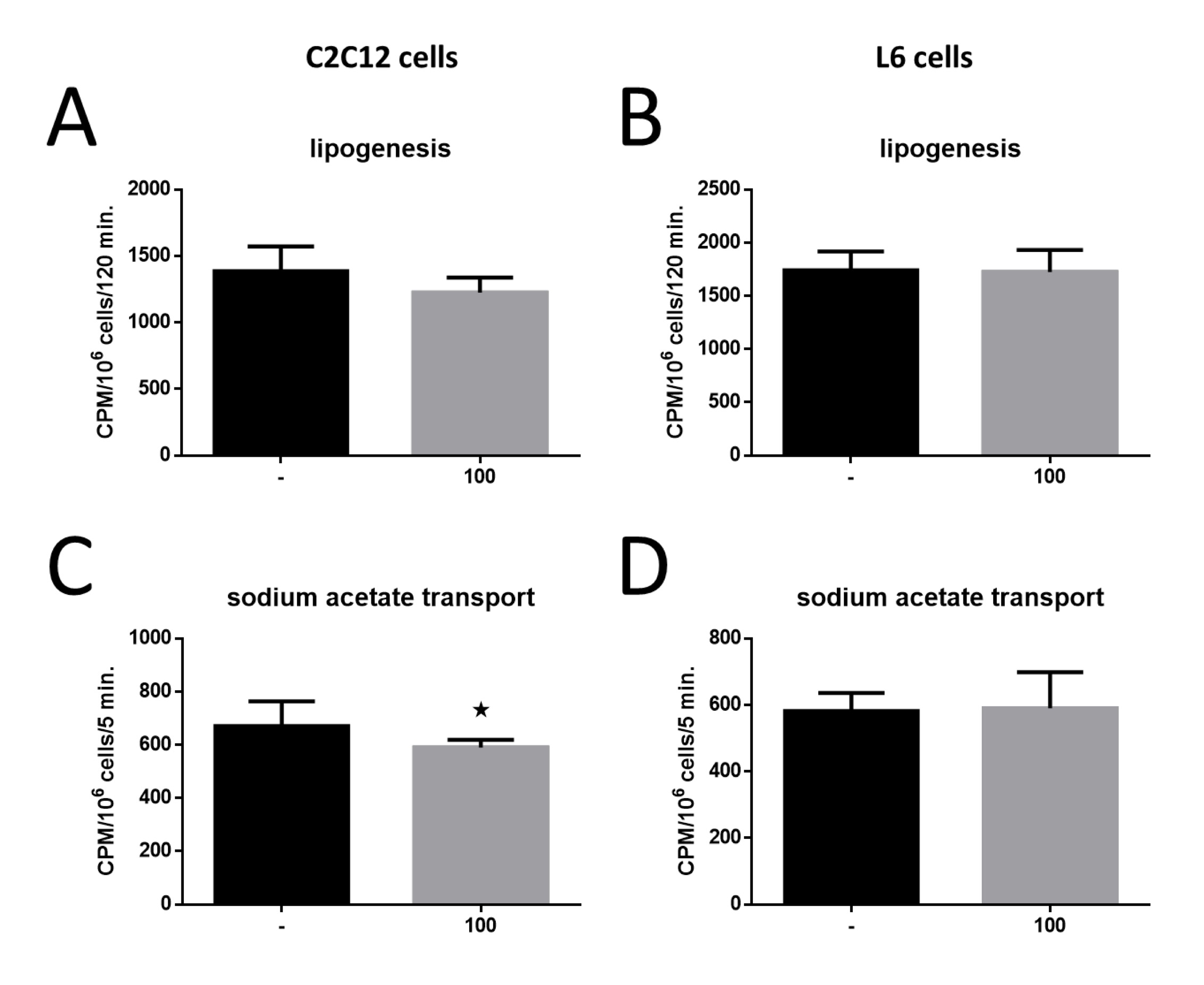
Fig. 7: Effect of MOTS-c on intensity of lipogenesis in C2C12 cell (A) and L6 cells (B) and sodium acetate transport to the C2C12 cell (C) and L6 cells (D). The results are shown as mean±SEM. Statistically significant differences are presented as *p<0.05 versus the corresponding control group using a one-way analysis of variance followed by Dunnett’s post hoc test.
Effects of MOTS-c on genes related to metabolic activity
Analyzing metabolic activity genes in response to MOTS-c treatment revealed significant differences between the two cell lines. In C2C12 cells, there was a notable increase in peroxisome proliferator-activated receptor γ (PPARγ) gene expression following MOTS-c treatment (*p<0, 05; Fig. 8A). In contrast, L6 cells demonstrated a significant decrease in PPARγ expression (*p< 0, 01; Fig. 8B), suggesting that MOTS-c may influence adipogenic pathways differently in these cell lines.
Regarding fatty acid binding protein 4 (FABP4) expression, C2C12 cells showed no significant changes following MOTS-c treatment (Fig. 8C). However, L6 cells exhibited a substantial reduction in FABP4 expression (*p<0, 05; Fig. 8D), highlighting the differential effects of MOTS-c on lipid metabolism-related genes.
Lastly, the glucose transporter-4 (GluT4) gene expression analysis revealed no significant changes in either C2C12 or L6 cells following MOTS-c treatment (Figures 8E and 8F). This lack of effect further illustrates the specificity of MOTS-c’s influence on metabolic activity genes across the different cell lines.
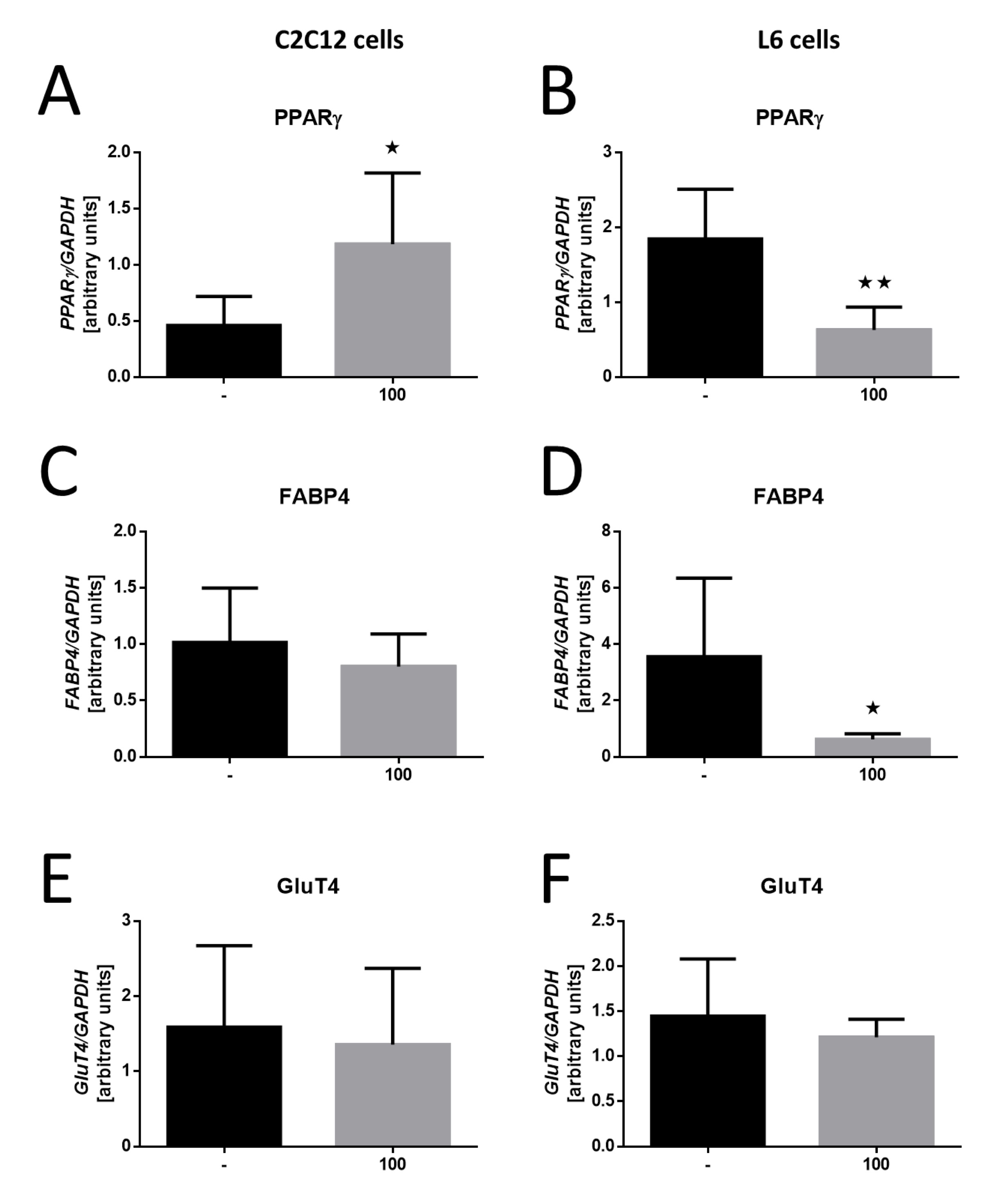
Fig. 8: Effect of MOTS-c on the expression of genes involved in lipid metabolism. Expression-related changes in PPARγ in C2C12 cell (A), PPARγ in L6 cells (B), FABP4 in C2C12 cells (C), FABP4 in L6 cells (D), GluT4 in C2C12 cells (E) and GluT4 in L6 cells (F). The results are shown as mean±SEM. Statistically significant differences are represented as *p<0.05 and **p<0.01 versus the corresponding control group using a one-way analysis of variance followed by Dunnett’s post hoc test.
Discussion
MOTS-c is a peptide intricately involved in energy metabolism, inherently linking it to tissues central to glucose and fatty acid metabolism. Given that muscle tissue constitutes the primary energy consumer in the body, our focus turned toward exploring the potential effect of MOTS-c on specific aspects of cellular function within this tissue. To this end, we used two widely used cell lines: murine C2C12 cells and rat L6 cells. The former belongs to the so-called white fibers (fast-twitch) that preferentially use glucose as their energy source, while the latter belongs to the so-called red fibers (slow-twitch), favoring fatty acids as their energy substrate [13]. Consequently, these cells manifest two radically distinct metabolic fiber types. Muscles with a predominance of fibers that primarily use glycolytic metabolism (type 2 fibers) are adapted to perform short contractions and rapid work, whereas oxidative metabolism-dominated muscles (type 1 fibers) excel at sustained contractions and gradual exertion [14, 15]. Other features that distinguish muscles with a predominance of white or red fibers are rapid fatigue, high ATPase activity, fewer mitochondria, myoglobin, respiratory enzymes, and glycogen, coupled with anaerobic activity. Conversely, red fibers exhibit the opposite attributes—reduced fatigue, lower ATPase activity, more mitochondria, myoglobin, respiratory enzymes, and glycogen, along with aerobic effect in red muscles [6, 22]. Given these metabolic and structural differences in the cell lines under study, we hypothesized that the impact of MOTS-c is different and would vary depending on the muscle fiber type, leading us to execute all experiments performed in parallel on both cell lines.
Our initial investigation revolved around the effect of MOTS-c on muscle cell survival using the MTT assay. Results unveil that MOTS-c significantly influences cell survival, albeit exclusively in the C2C12 line, with no discernible impact on the L6 line. Interestingly, our findings also suggest that MOTS-c triggers one of the pivotal pathways in the receptor signaling cascade—ERK kinase phosphorylation. This pathway is a key player in proliferation and differentiation processes [16, 17]. In L6 cells, this effect does not occur at all. To ensure that the effect of increasing survival is not related to cell proliferation, we evaluated the PCNA proliferation marker, revealing decreased expression of this protein. Integrating these findings, we can deduce that MOTS-c enhances cell survival not by stimulating cell proliferation but by extending the lifespan of individual cells [18]. These outcomes lend credence to the findings of prior studies, indicating the ability of MOTS-c to prolong cell lifespan [19].
Delineating the processes of proliferation and differentiation as opposing phenomena prompted our subsequent exploration: the impact of MOTS-c on the differentiation of C2C12 and L6 cells. These assessments were conducted on the 2nd and 6th days of differentiation, allowing observation of changes in factor expression at distinct stages. Our research focused on the myoD and myogenin genes, integral components of MRFs that orchestrate the determination and differentiation of skeletal muscle cells during embryogenesis and postnatal myogenesis [20]. We scrutinized gene expression at both RNA and protein levels while exclusively assessing the RNA levels of genes encoding myosin, specifically Mhc4, and Myh3, due to protein product size. Myosin, a hexameric protein pivotal to movement, exclusively surfaces in mature, fully-differentiated muscle cells. Our findings unveil MOTS-c’s differential effect on C2C12 cells on both the 2nd and 6th days of differentiation. Augmented expression of myoD and myogenin genes was confirmed at both RNA and protein levels. Furthermore, increased myosin gene expression is observed on the sixth day of differentiation, signifying progression in the process. Conversely, such pronounced effects are nearly absent in the L6 line. Only on the second day of differentiation is an increase in myoD and myosin gene expression noted, without concomitant changes in protein quantities. Particularly noteworthy is the observation that on the sixth day of differentiation, MOTS-c does not elicit any changes in differentiation markers for L6 cells. This underlines the insensitivity of L6 cells, which primarily metabolize fatty acids, to MOTS-c-induced differentiation stimulation. By contrast, C2C12 cells, avid for glucose-derived energy, show marked susceptibility to this factor. This indicates that MOTS-c is more engaged in glucose regulation than fatty acid metabolism, potentially holding a more profound effect on pancreatic function than adipose tissue. Notably, MOTS-c is not a strictly anabolic protein; while it can alleviate atrophy caused by diabetes and palmitic acid in vitro [21], it showcases versatility in its effects. Our findings align with García-Benlloch et al. study, indicating the promoting effect of MOTS-on C2C12 cell differentiation through the IL-6/JAK/STAT3 pathway [22].
The ongoing investigation into the role of MOTS-c in energy processes has unveiled its ability to curtail fat accumulation in white adipose tissue [23] and improve insulin sensitivity [24, 25]. Given muscles’ strong ties to insulin signaling and their high energy requirements, we enriched our study by subjecting cells to fatty acids to gauge their response to MOTS-c stimulation. Research has consistently identified fat deposition in tissues beyond the adipose tissue, particularly muscles [26], as a driver of insulin resistance and type 2 diabetes progression. Our findings indicate reduced lipid droplet accumulation in C2C12 cells under MOTS-c influence and, conversely, amplified accumulation in the L6 cell line. This juxtaposition can be attributed to the L6 cells’ preference for fatty acids as an energy source, coupled with a diverse effect of MOTS-c on these distinct cell lines. The lower accumulation in C2C12 cells might result from escalated catabolism of both glucose and fatty acids, while the heightened accumulation in L6 cells signifies increased anabolism, reflected in elevated lipid droplet storage.
As a result of such observations, the next step was to investigate the ability of muscle cells to synthesize fatty acids de novo in lipogenesis. The precursor of lipogenesis in this experiment was sodium acetate, a better substrate in this process than glucose, which, after entering the cells, can also participate in other metabolic processes [27]. We did not observe any changes in the intensity of the lipogenesis process after using MOTS-c in the experimental medium. This means that MOTS-c does not affect the ability of cells to synthesize fatty acids and triglycerides endogenously, but only their ability to accumulate lipids already synthesized.
A complementary result for the determination of lipogenesis was the determination of the intensity of intracellular transport of sodium acetate as a potential substrate for further lipid synthesis in cells. In this case, MOTS-c also had no stimulatory effect on the intracellular transport of sodium acetate in L6 cells. Moreover, for C2C12 cells, a decrease in transport intensity was observed after treating the cells with the MOTS-c peptide. This result additionally confirms that MOTS-c does not stimulate endogenous lipid synthesis in muscle cells, and their possible steatosis results from the absorption of lipids synthesized in other types of cells.
In the last step, the expression of several essential genes involved in carbohydrate-lipid metabolism was analyzed in both cell lines. In the case of peroxisome proliferator-activated receptor γ (PPARγ), an opposite effect of MOTS-c was observed on C2C12 and L6 cell lines. Once again, this proves the different metabolic nature of these lines. C2C12 cells prefer glucose metabolism, and the PPARγ receptor is involved, among others, in developing mitochondria, cell viability, and insulin resistance. Therefore, it is unsurprising that its expression increased in this cell line with a simultaneous decrease in the L6 cell line, which prefers fatty acid metabolism. It is worth noting that MOTS-c also acts through the PPARγ pathway in other body cells [28].
It is, therefore, not surprising that there are no changes in the expression of the fatty acid binding protein 4 (FABP4) gene in the C2C12 line. The decrease in the expression of this gene in the case of the L6 line is slightly more surprising, but remember that, as our results indicate, MOTS-c has practically no activating effect on the metabolism of these cells at any level.
Ultimately, it was also demonstrated that there were no changes in the expression of the glucose transporter 4 (GluT4) gene in both tested lines after treatment of cells with the MOTS-c peptide. Considering the current results, changes in expression could be expected in the C2C12 line. Still, the possible translocation of the Glut4 receptor to the cell membrane under the influence of MOTS-c without changing its amount in the cell remains an open question [29].
Conclusion
In conclusion, our study presents a distinctly different effect of MOTS-c on muscle cells, which depends on the metabolic fiber type they embody. MOTS-c significantly influences the physiology of C2C12 cells. It is worth noting that while our studies are conducted in vitro , they serve as intriguing precursors to forthcoming in vivo analyses. The significance of MOTS-c research is magnified by its potential use among athletes to augment performance [30]. Although doping control authorities have proposed a test to detect MOTS-c in plasma samples, the substance remains accessible on the black market. 2025 MOTS-c will be included in the World Anti-Doping Agency’s (WADA) List of Prohibited Substances and Methods [31]. However, in the future, after further in-depth research, MOTS-c would be used for therapeutic purposes in people with metabolic disorders.
Acknowledgements
Author Contributions
N.L.: conceptualization, methodology, investigation, writing original draft. E.P.O., P.K., D.S.: methodology & investigation. L.N. supervision. M.S.: writing, review & editing, project administration, funding acquisition, supervision.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Funding Sources
This research was funded by National Science Centre, grant no. 2018/31/D/NZ4/01121.
The publication was financed by the Polish Minister of Science and Higher Education as part of the Strategy of the Poznan University of Life Sciences for 2024-2026 in the field of improving scientific research and development work in priority research areas.
Disclosure Statement
The authors have no conflicts of interest to declare.
Disclosure of AI Assistance
The authors declare that they did not use AI tools at any stage when writing the work.
References
| 1 | Cheng W, Wang L, Yang B, Zhang R, Yao C, He L, Liu Z, Du P, Hammache K, Wen J, Li H, Xu Q, Hua Z: Self-renewal and differetiation of muscle satellite cells are regulated by the fas-associated death domain. J Biol Chem 2014;289:5040-50.
https://doi.org/10.1074/jbc.M113.533448 |
| 2 | Asfour HA, Allouh MZ, Said RS: Myogenic regulatory factors: The orchestrators of myogenesis after 30 years of discovery. Exp Biol Med 2018;243:118-28.
https://doi.org/10.1177/1535370217749494 |
| 3 | Zammit PS: Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol 2017;72:19-32.
https://doi.org/10.1016/j.semcdb.2017.11.011 |
| 4 | Wagatsuma A, Sakuma K: Mitochondria as a potential regulator of myogenesis. ScientificWorldJournal 2013;2013:593267.
https://doi.org/10.1155/2013/593267 |
| 5 | Merry TL, Chan A, Woodhead JST, Reynolds JC, Kumagai H, Kim SJ, Lee C: Mitochondrial-derived peptides in energy metabolism. Am J Physiol Endocrinol Metab 2020;319:E659-66.
https://doi.org/10.1152/ajpendo.00249.2020 |
| 6 | Zheng Y, Wei Z, Wang T MOTS-c: A promising mitochondrial-derived peptide for therapeutic exploitation. Front Endocrinol (Lausanne) 2023;14:1-6.
https://doi.org/10.3389/fendo.2023.1120533 |
| 7 | Kołodziejski PA, Pruszyńska-Oszmałek E, Wojciechowicz T, Sassek M, Leciejewska N, Jasaszwili M, Billert M, Małek E, Szczepankiewicz D, Misiewicz-Mielnik M, Hertig I, Nogowski L, Nowak KW, Strowski MZ, Skrzypski M: The role of peptide hormones discovered in the 21st century in the regulation of adipose tissue functions. Genes (Basel) 2021;12:756.
https://doi.org/10.3390/genes12050756 |
| 8 | Bhullar KS, Shang N, Kerek E, Wu K, Wu J: Mitofusion is required for MOTS‐c induced GLUT4 translocation. Sci Rep 2021;11:1-12.
https://doi.org/10.1038/s41598-021-93735-2 |
| 9 | Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P: The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 2015 Mar 3;21(3):443-54.
https://doi.org/10.1016/j.cmet.2015.02.009 |
| 10 | Kumagai H, Coelho AR, Wan J, Mehta HH, Yen K, Huang A, Zempo H, Fuku N, Maeda S, Oliveira PJ, Cohen P, Kim S: MOTS-c reduces myostatin and muscle atrophy signaling. Am J Physiol Endocrinol Metab 2021;320:E680-90.
https://doi.org/10.1152/ajpendo.00275.2020 |
| 11 | Lawson MA, Purslow PP: Differentiation of myoblasts in serum-free media: Effects of modified media are cell line-specific. Cells Tissues Organs 2000;167:130-7.
https://doi.org/10.1159/000016776 |
| 12 | Chiu YJ, Lo YH, Yang JS, Kuo SC, Tsai SC: Curcumin derivative MTH-3 regulates palmitate-induced insulin resistance in mouse myoblast C2C12 cells. In vivo (Brooklyn) 2021;35:3181-91.
https://doi.org/10.21873/invivo.12613 |
| 13 | Abdelmoez AM, Puig LS, Smith JAB, Gabriel BM, Savikj M, Dollet L, Chibalin AV, Krook A, Zierath JR, Pillon NJ: Comparative profiling of skeletal muscle models reveals heterogeneity of transcriptome and metabolism. Am J Physiol Cell Physiol 2020;318:C615-26.
https://doi.org/10.1152/ajpcell.00540.2019 |
| 14 | Schiaffino S, Reggiani C: Fiber types in Mammalian skeletal muscles. Physiol Rev 2011;91:1447-531.
https://doi.org/10.1152/physrev.00031.2010 |
| 15 | Wilson JM, Loenneke JP, Jo E, Wilson GJ, Zourdos MC, Kim JS: The effects of endurance, strength, and power training on muscle fiber type shifting. J Strength Cond Res 2012;26:1724-9.
https://doi.org/10.1519/JSC.0b013e318234eb6f |
| 16 | Mebratu Y, Tesfaigzi Y: How ERK1/2 activation controls cell proliferation and cell death is subcellular localization the answer? Cell Cycle 2009;8:1168-75.
https://doi.org/10.4161/cc.8.8.8147 |
| 17 | Michailovici I, Harrington HA, Azogui HH, Yahalom-Ronen Y, Plotnikov A, Ching S, Stumpf MP, Klein OD, Seger R, Tzahor E: Nuclear to cytoplasmic shuttling of ERK promotes differentiation of muscle stem/progenitor cells. Development (Cambridge) 2014;141:2611-20.
https://doi.org/10.1242/dev.107078 |
| 18 | Wan W, Zhang L, Lin Y, Rao X, Wang X, Hua F, Yinf J: Mitochondria-derived peptide MOTS-c: effects and mechanisms related to stress, metabolism and aging. J Transl Med 2023;21:1-13.
https://doi.org/10.1186/s12967-023-03885-2 |
| 19 | Fuku N, Pareja-Galeano H, Zempo H, Alis R, Arai Y, Lucia A, Hirose N: The mitochondrial-derived peptide MOTS-c: A player in exceptional longevity? Aging Cell 2015;14:921-3.
https://doi.org/10.1111/acel.12389 |
| 20 | Londhe P, Davie JK: Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet Muscle 2011;1:14.
https://doi.org/10.1186/2044-5040-1-14 |
| 21 | Kumagai H, Coelho AR, Wan J, Mehta HH, Yen K, Huang A, Zempo H, Fuku N, Maeda S, Oliveira PJ, Cohen P, Kim S: MOTS-c reduces myostatin and muscle atrophy signaling. Am J Physiol Endocrinol Metab 2021;320:E680-90.
https://doi.org/10.1152/ajpendo.00275.2020 |
| 22 | García-Benlloch S, Revert-Ros F, Blesa JR, Alis R: MOTS-c promotes muscle differentiation in vitro. Peptides (NY) 2022;155:170840.
https://doi.org/10.1016/j.peptides.2022.170840 |
| 23 | Kim SJ, Miller B, Mehta HH, Xiao J, Wan J, Arpawong TE, Yen K, Cohen P: The mitochondrial-derived peptide MOTS-c is a regulator of plasma metabolites and enhances insulin sensitivity. Physiol Rep 2019;7:1-12.
https://doi.org/10.14814/phy2.14171 |
| 24 | Lu H, Wei M, Zhai Y, Li Q, Ye Z, Wang L, Luo W, Chen J, Lu Z: MOTS-c peptide regulates adipose homeostasis to prevent ovariectomy-induced metabolic dysfunction. J Mol Med 2019;97:473-85.
https://doi.org/10.1007/s00109-018-01738-w |
| 25 | Wan W, Zhang L, Lin Y, Rao X, Wang X, Hua F, Ying J: Mitochondria-derived peptide MOTS-c: effects and mechanisms related to stress, metabolism and aging. J Transl Med 2023;21:1-13.
https://doi.org/10.1186/s12967-023-03885-2 |
| 26 | Hana CA, Klebermass EM, Balber T, Mitterhauser M, Quint R, Hirtl Y, Klimpke A, Somloi S, Hutz J, Sperr E, Eder P, Jašprová J, Valášková P, Vítek L, Heiss E, Wagner KH: Inhibition of Lipid Accumulation in Skeletal Muscle and Liver Cells: A Protective Mechanism of Bilirubin Against Diabetes Mellitus Type 2 Front Pharmacol 2021;11:1-14.
https://doi.org/10.3389/fphar.2020.636533 |
| 27 | Karvinen S, Fachada V, Sahinaho U-M, Pekkala S, Lautaoja JH, Mäntyselkä S, Permi P, Hulmi JJ, Silvennoinen M, Kainulainen H: Branched-Chain Amino Acid Deprivation Decreases Lipid Oxidation and Lipogenesis in C2C12 Myotubes. Metabolites 2022;12:328.
https://doi.org/10.3390/metabo12040328 |
| 28 | Lu P, Li X, Li B, Li X, Wang C, Liu Z, Ji Y, Wang X, Wen Z, Fan J, Yi C, Song M, Wang X: The mitochondrial-derived peptide MOTS-c suppresses ferroptosis and alleviates acute lung injury induced by myocardial ischemia reperfusion via PPARγ signaling pathway. Eur J Pharmacol 2023;953:175835.
https://doi.org/10.1016/j.ejphar.2023.175835 |
| 29 | Bhullar KS, Shang N, Kerek E, Wu K, Wu J: Mitofusion is required for MOTS‐c induced GLUT4 translocation. Sci Rep 2021;11:14291.
https://doi.org/10.1038/s41598-021-93735-2 |
| 30 | Knoop A, Thomas A, Thevis M: Development of a mass spectrometry based detection method for the mitochondrion-derived peptide MOTS-c in plasma samples for doping control purposes. Rapid Communications in Mass Spectrometry 2019;33:371-80.
https://doi.org/10.1002/rcm.8337 |
| 31 | WORLD ANTI-DOPING CODE INTERNATIONAL STANDARD PROHIBITED LIST 2025 [2024 Nov 24]
https://doi.org/10.46607/iamj12092024 www.wada-ama.org/sites/default/files/2024-09/2025list_en_final_clean_12_september_2024.pdf |