Significant Association of Candidate Genes (AGTR1 and TGF-Β1) Polymorphism with Diabetic Nephropathy in Diabetes Mellitus Type 2 Patients
bDepartment of Zoology, Islamia College Peshawar,
cZoology Department, College of Science, King Saud University, P.O. Box: 2455, 11451, Riyadh, Saudi Arabia,
dCentre for Applied Mathematics and Bioinformatics (CAMB), Gulf University for Science and Technology, Hawally, Kuwait,
eDepartment of RNA Sciences, The Brain Institute of America, New Haven, CT, USA
Keywords
Abstract
Background/Aims:
Diabetic nephropathy (DN) is one of the complications of diabetes mellitus (DM). This study aimed to investigate the association between genetic polymorphisms, specifically AGTR1 (rs5186) and TGF-β1 (rs1800470), and the risk of developing Diabetic nephropathy (DN) in type 2 diabetes mellitus patients, compared to those without DN and healthy controls.Methods:
A case-control study was conducted on 165 diabetic patients (59 with diabetic nephropathy (DN) and 54 without DN (DM)), and 52 healthy controls (HC). The genotyping was done using amplification refractory mutation system method (ARMS-PCR). Age, gender, and duration of diabetes were matched across groups. Clinical parameters including FBS, RBS, HbA1C, creatinine, urea, SBP, DBP, total cholesterol, triglycerides, LDL, and BMI were assessed.Results:
Diabetic patients with nephropathy exhibited significantly higher levels of clinical parameters compared to those without nephropathy and healthy controls. The risk allele of AGTR1 , C (p <0.0001), and risk allele containing genotypes AC (p <0.0001) and CC (p - 0.0010) were significantly higher in DN patients compared to DM and HC groups. Similarly, the TGF-β1 risk allele C (p - 0.0001), and corresponding genotypes TC (p - 0.0038) and CC (p - 0.0027) were significantly associated with increased risk of diabetic nephropathy compared to DM and HC groups.Conclusion:
The data showed significant association of AGTR1 (rs5186) and TGF-β1 (rs1800470) polymorphism with an increased risk of diabetic nephropathy in type 2 diabetes mellitus patients. More investigation will be required to disseminate the results, while increasing the samples size and using whole genome sequencing.Introduction
Diabetes mellitus (DM) is a complex disorder, which is very common and rapidly growing all over the world [1]. In 2002, about 173 million cases of DM were estimated worldwide and this number was predicted to increase by 350 million in 2030 [2]. The disease is influenced by both environmental and genetic factors [3]. Diabetic Nephropathy (DN) is one of the microvascular complications of DM. It is estimated that approximately 30–40% of all patients with diabetes develop DN and is the leading cause of end stage renal disease (ESRD) [4]. The ESRD is considered the leading cause of kidney failure and death in patients with DM and that is why it is important to prevent diabetes development into DN [5]. Hyperglycemia, hyperlipidemia, hypertension, advanced glycation products accumulation, duration of diabetes, familial clustering and genetic determinants are some of the risk factors which can make a diabetic patient susceptible to DN [2].
Transforming growth factor beta (TGF-β ) belongs to a group of multifunctional growth factors which control various biological processes such as apoptosis, senescence, healing of wounds, tumor metastasis and suppression, cell division, differentiation and immunity [6]. In humans, there are three isoforms of TGF-β namely TGF-β1 , TGF-β2 and TGF-β3 . Among these the TGF-β1 is the most abundant and is highly conserved in primary sequence through evolution [7]. Since the TGF-β1 controls the production and degradation of the renal extracellular matrix (ECM), as well as the expression of cell adhesion molecule receptors, it is said that it might be directly involved in developing a kidney disease [8]. The high levels of glucose in blood activates TGF-β1 transcription in mesangial cells. Such high glucose levels and the recombinant TGF-β increase the synthesis of ECM in the renal cells and also participate in cell hypertrophy [9]. The human TGF-β1 gene is located on chromosome 19q13.1–13.3 [10] and more than 10 polymorphic loci are currently known, which are distributed in exons and introns regions as well as in the 5’- flanking region [3]. The polymorphisms at codons 10 and 25 are reported to be associated with increased or decreased levels of TGF-β1 production in vitro [11]. This increase or decrease in the production of TGF-β1 has been linked to various diseases such as atherosclerosis and fibrotic diseases of the liver, kidney and lungs [12]. Various SNPs are present in certain loci of the TGF-β1 gene which affect its regulation and expression levels. Among such SNPs our focus in this study was on the TGF-β1 gene polymorphism T > C (rs1800470).
Another candidate gene, Angiotensin II Receptor Type 1 (AGTR1 ) is found to be a highly polymorphic but the rs5186 (A1166C) polymorphism is greatly evaluated [13]. It has been investigated in different populations for its association with the risk and susceptibility to develop DN. Angiotensin II is one of the main enzymes of the renin-angiotensin aldosterone system (RAAS) which plays an important role in maintaining the blood pressure [14]. Hyperglycemia, which could activate RAAS, increases tissue angiotensin II that induces glomerular hyperfiltration, oxidative stress, thrombosis, endothelial damage, inflammation and vascular remodeling [15]. Studies on Animal models have also revealed that RAAS may participate in transition of epithelial cells to mesenchymal cells and thereafter renal fibrosis under hyperglycemic environment. Such studies discovered that the angiotensin II and its receptor may be a potential predictor for developing DN in patients with diabetes [16]. It has been suggested that the mutation A1166C (rs5186) of AGTR1 gene is one of the potential candidate genes for DN. This A1166C variation is located in the 3′ end of the non-coding regions and it may affect the stability and translation of the mRNA [17].
However, GWAS have identified the selected SNPs in various ethnicities with inconsistent results. Understanding the clinical implications of these genetic variants is essential for improving risk prediction, patient stratification, and therapeutic targeting in individuals with type 2 diabetes mellitus. Therefore, our study aims to investigate the presence and association of TGF-β1 and AGTR1 gene polymorphisms with the risk of developing DN of diabetic type 2 patients of Khyber Pakhtunkhwa, Pakistan.
Materials and Methods
Study subjects, inclusion, exclusion criteria and Blood sampling
This case control study was performed between February and December 2020 in the Institute of Biotechnology and Genetic Engineering (Health Division), The University of Agriculture Peshawar. A total of 165 participants were enrolled in the study including 59 patients with diabetic nephropathy (DN), 54 patients without nephropathy (DM) and a control group consisting of 52 healthy controls (HC). A written informed consent was obtained from each participant after describing the aim of the study. T2DM was diagnosed on the basis of WHO criteria of fasting blood glucose level of ≥7.0mmol/L (126mg/dL). Subjects with >10 years of diabetes duration and having urinary albumin levels <30 μg/mg creatinine measured on two consecutive collections with no issues in the kidneys were placed in the DM group. The status of the DN was determined on the basis of clinical features, laboratory reports and questionnaires. Subjects with >10 years of diabetes duration with urinary albumin levels of ≥300μg/mg of creatinine in at least two of three fasting urine collections over a period of 3 months were placed in DN group. Subjects of the control group consisted of healthy participants without any history of diabetes and any kind of kidney disease. Exclusion criteria in this study were the subjects who did not belong to Khyber Pakhtunkhwa population, patients with Type 1 diabetes, duration of diabetes less than 10 years, urinary tract infection, pregnancy, smoking, hematuria, single kidney, kidney stones or any other causes of nephropathy except diabetes type 2. About 4 mL blood samples were taken in EDTA tubes for molecular study keeping in notice the standard biosafety protocol.
DNA isolation and PCR confirmation
All the blood samples were processed for genomic DNA extraction using the non-enzymatic/salting out method adopted in our lab [18, 19]. For SNPs genotyping, amplification refractory mutation system (ARMS-PCR) was carried out. NCBI Primer-BLAST software was used for designing primers (Table 1). The PCR mixture of 10μL was prepared consisting of 5μL Dream Taq Green master mix (Thermo Fischer), 3μL of ddH2O, 1μL of template DNA, and 0.5μL of each forward and reverse primer. The PCR amplification conditions for the AGTR1 variant were; initial denaturation at 95° C for 5 min, proceeding with 32 cycles of denaturation at 95° C for 30 s, annealing at 58° C for 30 s, extension at 72° C for 30 s, and final extension at 72° C for 5 min. The PCR amplification conditions for the TGF -β1 codon 1 (T869C) were; 95°C for 1 min followed by 30 cycles of 95°C for 15s, 65°C for 30s, and 72°C for 40s, and final extension at 72° C for 5 min. The amplified PCR products were analyzed on 2% agarose gel.
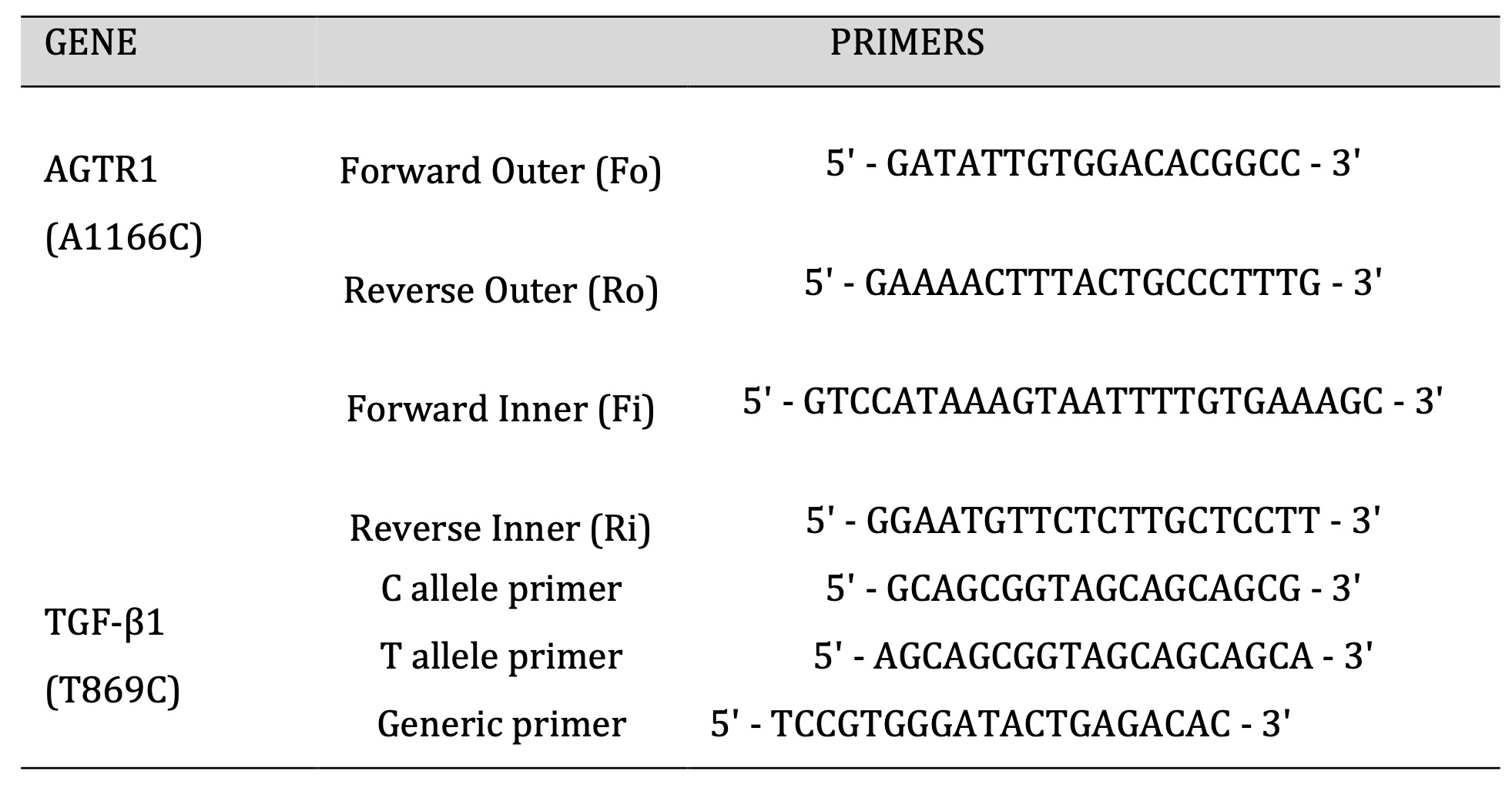
Table 1: Primers used for AGTR1 (A1166C) and TGF-β1 (T869C).
Statistical analysis
Statistical analysis was performed using SPSS software version 2.0. Descriptive statistical method was used for demographic data analysis with P < 0.05 was considered significant. The data is presented as Mean ± SD. For categorical data, the Pearson Chi Square test was used. Deviations from Hardy–Weinberg equilibrium was tested. Comparison among different study groups were performed using one-way ANOVA. Genotypic and allelic frequencies were calculated. The comparison of genotypes was performed by obtaining Odd ratios and 95% Confidence interval values using the online software called Medcalc Odd ratio calculator. The P > 0.05 was considered non-significant.
Results
Demographic and clinical characteristics of DN, DM and HC group
A total of 165 subjects containing 59 DN, 54 DM and 52 HC were recruited in this study. Demographic and clinical characteristics of the study subjects is summarized in Table 2. Comparison of DN group with DM and HC showed a significantly higher levels of FBS (P <0.001), RBS (P <0.001), HbA1c (P <0.001), Creatinine (P <0.001), Urea (P = <0.001), Systolic blood pressure (P <0.001), Diastolic blood pressure (P <0.001), Total cholesterol (P <0.001), Triglycerides (P <0.001), LDL (P =0.007), HDL (P =0.054), Hypertension (P =0.023), and BMI (P =0.011). Furthermore, the eGFR was extremely lower in the DN patients than DM and HC groups. The age, gender and duration of the diabetes showed no significant difference among the study groups (Table 2).
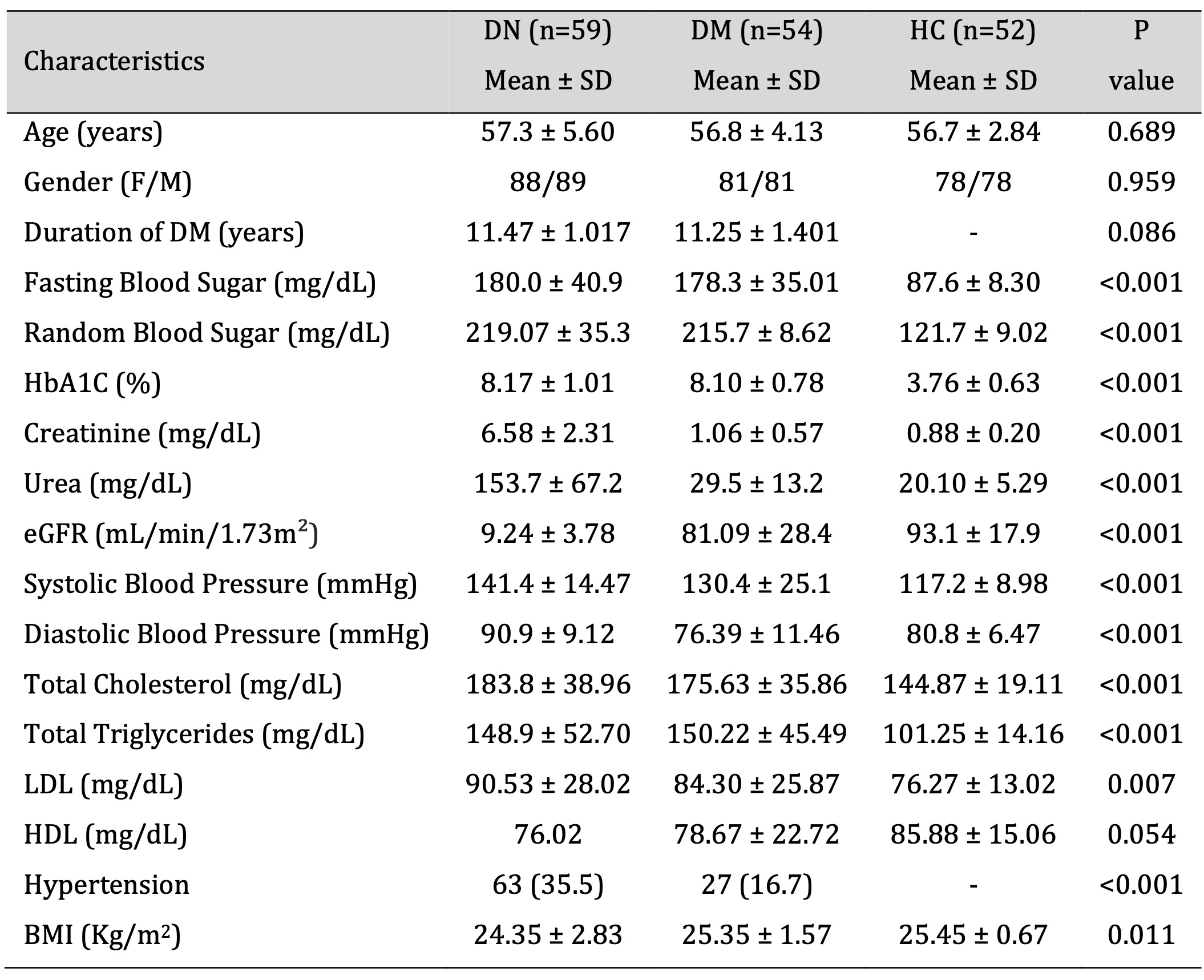
Table 2: Demographics and biochemical characteristics of DN, DM and HC group. Data presented in Mean ± Standard deviation. P value calculated through chi square test. DN - diabetic nephropathy, DM – Diabetes mellitus, HC - Healthy control, eGFR – estimated Glomerular Filtration Rate, LDL - low density lipids, HDL - high density lipids, BMI - Body mass index.
Allelic and genotypic frequencies of TGF-β1 (rs1800470) and AGTR1 (rs5186) in the study groups
The allelic and genotypic frequencies of AGTR1 (rs5186) and TGF-β1 (rs1800470) was confirmed using ARMS-PCR, run on 2% agarose gel compared with 1KB DNA ladder (Fig. 1). Our results indicated that the AGTR1 (rs5186) risk allele C and risk allele containing genotypes AC and CC showed significant association with the increase risk of diabetic nephropathy compared with DM and HC groups (DN vs. HC, C- p =<0.0001 AC-p =<0.0001, CC- p =0.0010; DN vs. DM, C- p =0.0001, AC- p =0.0003, CC- p =0.0022; DM vs. HC, C- p =0.2055, AC- p =0.1182, CC- p =0.8673) (Table 3). Similarly, the TGF-β1 (rs1800470) risk allele C and risk allele containing genotypes TC and CC were significantly associated with increased risk of diabetic nephropathy (DN vs. HC, C- p =<0.0001, TC-p =<0.0038, CC- p =0.0027; DN vs. DM, C- p =0.0123, TC- p =0.0328, CC- p =0.0282; DM vs. HC, C- p =0.1415, TC- p =0.2983, CC- p =0.2903) (Table 3).
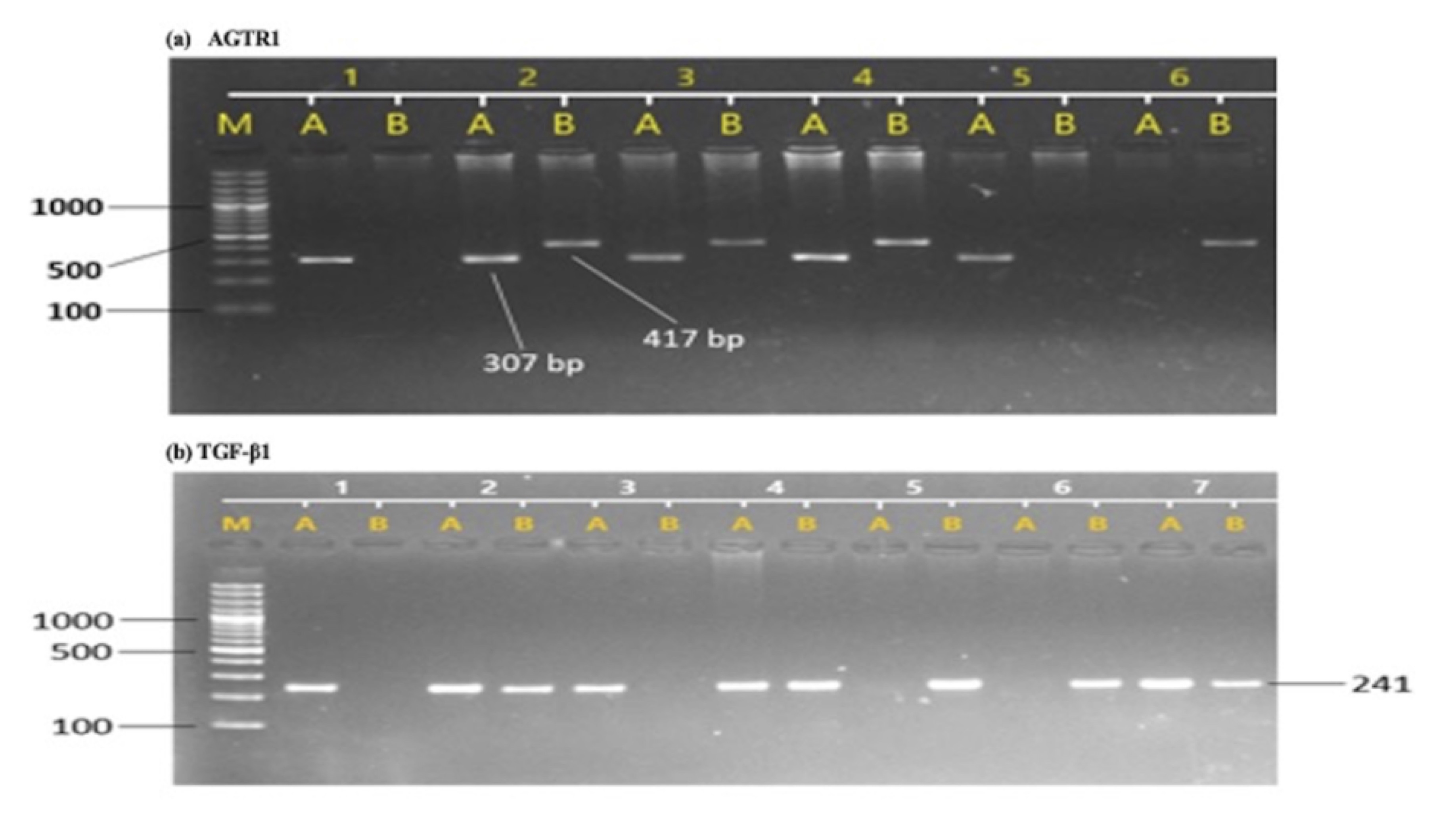
Fig. 1: Sample gel pictures of T869C of TGF-β1 and A1166C of AGTR1 gene polymorphism. (a), AGTR1 polymorphism, M represents DNA ladder, A represent wild allele and B represents mutant allele, sample 1 represents AA genotype, sample 2 represents AC genotype and samples 6 represents CC genotype. (b), TGF-β1 polymorphism, M represents DNA ladder, A represent wild allele and B represents mutant allele, sample 1 represents TT genotype, samples 2 represents TC genotype and samples 5 represents CC genotype.
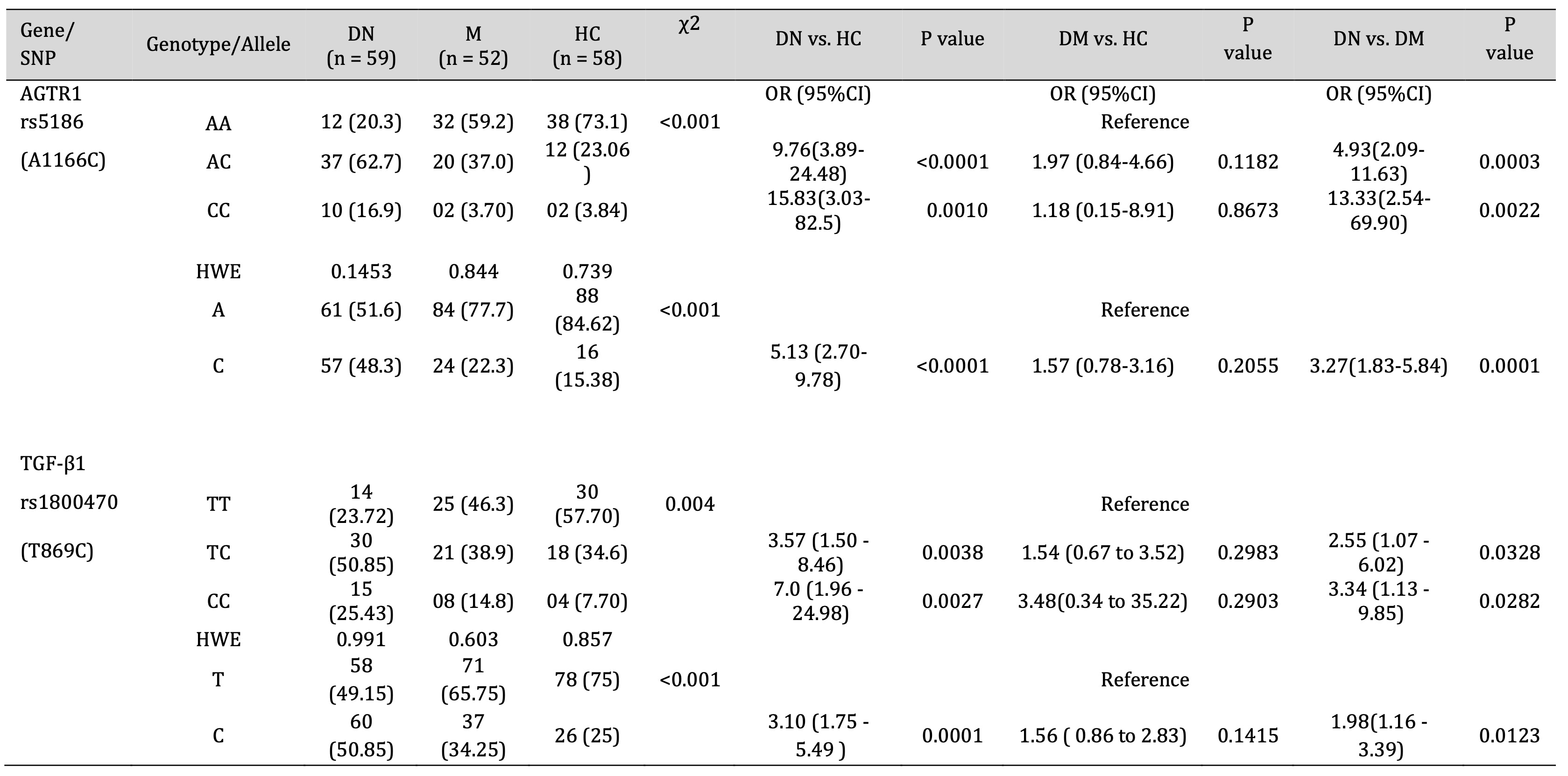
Table 3: Association of AGTR1 and TGF-β1 Gene polymorphism with DN, DM and HC group. HWE - Hardy Weinberg equilibrium, OR - Odd ratios, CI - Confidence interval, χ2 - Chi square value.
Discussion
TGF-β is said to be an important regulator in the production and degradation of the renal extracellular matrix [9]. When it is overexpressed, it could cause tissue fibrosis which eventually leads to failure of the organ [20]. Extreme accumulation of extra cellular matrix and the renal hypertrophy could occur because of higher concentration of TGF-β1 , such changes progress to DN [21]. Diabetic patients were found to have an elevated renal production of TGF-β protein [9]. Some other factors that are known to be involved in the initiation and progression to DN are excessively increased blood sugar levels, renin angiotensin system activation, increased intraglomerular pressure and hypertension, which are said to induce the production of TGF-β in the kidneys [22].
The actual mechanism to know how exactly the gene polymorphism in TGF-β1 affect the susceptibility to DN is not very clear yet. It has been known that the gene TGF-β1 is located on the chromosome 19 (q13.1-13.3) and has a total of 7 exons and 6 introns [23]. Various polymorphisms have been identified in the TGF-β1 gene [24]. One of them is the rs1800470, which occurs in exon number 1, at position 869. Here in this study, we have investigated TGFβ-1 gene as a candidate gene for susceptibility to DN in patients with type 2 diabetes of Khyber Pakhtunkhwa, Pakistan. Our analysis indicated the association between allelic and genotypic frequencies of the variant rs1800470 and susceptibility to DN. The CC genotypes and C allele were more frequent in DN group as compared to DM and HC. The increase in TC genotype in patients with DN showed that it could be a risk factor of developing DN in individuals with prolonged history of diabetes. Our results are similarly with a case control study, which was carried out on Chinese patients who had a history of having diabetes mellitus type 2 for over 10 years. Their result suggested that the TGF-β1 gene polymorphism is associated with susceptibility to diabetic nephropathy [22]. Another similar study was carried out in patients with type 1 diabetes to see if it contributes to genetic predisposition to diabetic nephropathy [7]. A study on patients of Egyptian origin and their data showed that C allele and C allele containing genotypes might be susceptible and the T allele and TT genotypes may be protective factors [3]. These results were in accordance with our findings. However, some studies reported results which were in contrast with our study. Similar study conducted on type 2 diabetes patients, in order to see the association of the TGF-β1 gene polymorphism with diabetic nephropathy. Their results suggested that TGF-β1 gene polymorphism is not associated with the progression of diabetic nephropathy [25]. A study on German populations concluded that the T allele rather than C allele was associated with ESRD susceptibility in patients with type 2 diabetes [26]. However, they recommend more investigation will be required to figured out the exact association while using next generation sequencing with increase samples size.
Here, we have also investigated the association of AGTR1 (rs5186). The AGTR1 gene is said to be located on the 3q21-25, having a length of less than 55 kb. This angiotensin II receptor actually regulates the level of an enzyme angiotensin II, which has great important in RAAS. RAAS is a system that regulates vasoconstriction, sodium reabsorption and the inflammatory cascade, which is believed to put a positive effect on the development of DN. Several studies have been done on AGTR1 gene polymorphisms, especially on rs5186 [17]. Our results indicated the association of AGTR1 gene polymorphism with susceptibility to DN. We noted significantly higher level of risk allele C in the DN group as compared to those in DM and HC group. The AC genotype was also higher in DN group than DM and HC group. Similarly, a study from North Indian origin stated that C allele and CC genotype of AGTR1 gene was significantly higher in patients with DN and the patients with AC genotype and CC genotype were found to be associated with increased risk for developing DN as compared to patients with AA genotype [27]. Another study conducted and observed the association of the AGTR1 genotype with the development of renal disease and progression to ESRD [24]. A significant difference in the frequency of the C allele and CC genotypes between patients and control group was noted. However, a [28] case control study conducted by [29] found no significant difference in the frequency of AGT1R genotypes between the patients and the control group. Their study revealed a higher but not a significantly different frequency of AGT1R AC+CC genotype in patients as compared to controls. A Meta-Analysis study of was conducted on eight articles, four were conducted in patients of Caucasian origin, and four were in Asians. They did not find an association of AGTR1 gene polymorphism and ESRD risk in Caucasians and Asians [30]. Therefore, to disseminate the investigation more similar research will be required to figure out the exact association of the AGTR1 gene polymorphism with DN in our population.
Conclusion
In conclusion, our research suggested an association of allelic and genotypic frequencies of the variant rs5186 of AGTR1 and rs1800470 of TGF-β1 as an increased risk of diabetic nephropathy. However, one of the short comings of this study was the small sample size. These results should be confirmed in larger sample size as well as in different ethnic groups while using modern sequencing technologies.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R191), King Saud University, Riyadh, Saudi Arabia
Funding
There is no funding received to conduct the study, the authors contributed for the study.
Authors’ Contributions
MI did the experimental work and wrote the first draft under the supervision of NUK. MI, NA, NUK, MHA, IA and BDA help in data analysis and critically review the final manuscript.
Ethical approval and consent to participate
Ethical approval was taken from the Institute of Biotechnology and Genetic Engineering (IBGE), The University of Agriculture Peshawar, Pakistan under (IBGE, UAP/2021/002). A written informed consent was taken from all the participants after explaining the aim of the study.
Authors Declaration
All the authors approved the final manuscript and agreed to publish it.
Consent to publish
All the authors have read and approved the article for publication.
Availability of data and materials
All the necessary data are included in manuscript, related data will be provided on request from corresponding author.
Disclosure Statement
The authors declared no competing interests.
References
| 1 | Stumvoll M, Goldstein BJ, Haeften TW Van. Haeften_Pathogenesis of type 2 diabetes.pdf. Lancet 2005;365:1333-1346.
https://doi.org/10.1016/S0140-6736(05)61032-X |
| 2 | Mehrabzadeh M, Pasalar P, Karimi M, Abdollahi M, Daneshpour M, Asadolahpour E, et al. Association between ELMO1 gene polymorphisms and diabetic nephropathy in an Iranian population. J Diabetes Metab Disord 2016;15. DOI: 10.1186/s40200-016-0265-3
https://doi.org/10.1186/s40200-016-0265-3 |
| 3 | El-Sherbini SM, Shahen SM, Mosaad YM, Abdelgawad MS, Talaat RM. Gene polymorphism of transforming growth factor-β1 in Egyptian patients with type 2 diabetes and diabetic nephropathy. Acta Biochim Biophys Sin (Shanghai) 2013;45:330-388.
https://doi.org/10.1093/abbs/gmt003 |
| 4 | Collins AJ, Kasiske B, Herzog C, Chavers B, Foley R, Gilbertson D, et al. United States Renal Data System 2006 Annual Data Report Abstract. Am J Kidney Dis. 2007;49(SUPPL. 1). DOI: 10.1053/j.ajkd.2006.11.019
https://doi.org/10.1053/j.ajkd.2006.11.019 |
| 5 | Mou X, Liu Y, Zhou D, Hu Y, Ma G, Shou C, et al. Different risk indictors of diabetic nephropathy in transforming growth factor-beta1 T869c CC/CT genotype and TT genotype. Iran J Public Health 2016;45:761-767.
|
| 6 | Chang AS, Hathaway CK, Smithies O, Kakoki M. Transforming growth factor-β1 and diabetic nephropathy. Am J Physiol - Ren Physiol 2016;310:689-696.
https://doi.org/10.1152/ajprenal.00502.2015 |
| 7 | Patel A, Scott WR, Lympany PA, Rippin JD, Gill G V., Barnett AH, et al. The TGF-β1 gene codon 10 polymorphism contributes to the genetic predisposition to nephropathy in Type 1 diabetes. Diabet Med. 2005;22(1):69-73.
https://doi.org/10.1111/j.1464-5491.2005.01376.x |
| 8 | McLennan S V., Fisher E, Martell SY, Death AK, Williams PF, Lyons JG, et al. Effects of glucose on matrix metalloproteinase and plasmin activities in mesangial cells: Possible role in diabetic nephropathy. Kidney Int Suppl. 2000;58(77). DOI: 10.1046/j.1523-1755.2000.07713.x
https://doi.org/10.1046/j.1523-1755.2000.07713.x |
| 9 | Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease: The case for transforming growth factor-β as a key mediator. Diabetes 1995;44:1139-1146.
https://doi.org/10.2337/diab.44.10.1139 |
| 10 | Fujii D, Brissenden JE, Derynck R, Francke U. Transforming growth factor β gene maps to human chromosome 19 long arm and to mouse chromosome 7. Somat Cell Mol Genet 1986;12:281-288.
https://doi.org/10.1007/BF01570787 |
| 11 | Awad MR, El-Gamel A, Hasleton P, Turner DM, Sinnott PJ, Hutchinson I V. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation 1998;66:1014-1020.
https://doi.org/10.1097/00007890-199810270-00009 |
| 12 | Blobe GC, Schiemann WP, Lodish HF. Role of Transforming Growth Factor β in Human Disease. N Engl J Med 2000 ;342:1350-1358.
https://doi.org/10.1056/NEJM200005043421807 |
| 13 | Halder K, Purkait P. Association of Angiotensin II Type I Receptor ( AGTR1 ) Gene Polymorphism and Type 2 Diabetes & Nephropathy among the Eastern Indian Bengali Patients. Diabetes Obes Int J 2020;5:1-13.
https://doi.org/10.23880/doij-16000224 |
| 14 | Putnam K, Shoemaker R, Yiannikouris F, Cassis LA. The renin-angiotensin system: A target of and contributor to dyslipidemias, altered glucose homeostasis, and hypertension of the metabolic syndrome. Am J Physiol Hear Circ Physiol 2012;302: 10.1152/ajpheart.00796.2011.
https://doi.org/10.1152/ajpheart.00796.2011 |
| 15 | Ruggenenti P, Bettinaglio P, Pinares F, Remuzzi G. Angiotensin converting enzyme insertion/deletion polymorphism and renoprotection in diabetic and nondiabetic nephropathies. Clin J Am Soc Nephrol 2008;3:1511-1525.
https://doi.org/10.2215/CJN.04140907 |
| 16 | Zhou L, Xue H, Yuan P, Ni J, Yu C, Huang Y, et al. Angiotensin AT1 receptor activation mediates high glucose-induced epithelial-mesenchymal transition in renal proximal tubular cells. Clin Exp Pharmacol Physiol 2010;37:152-157.
https://doi.org/10.1111/j.1440-1681.2010.05421.x |
| 17 | Wei L, Xiao Y, Li L, Xiong X, Han Y, Zhu X, et al. The Susceptibility Genes in Diabetic Nephropathy. Kidney Dis 2018;4:226-237.
https://doi.org/10.1159/000492633 |
| 18 | Adnan F, Khan NU, Iqbal A, Ali I, Petruzziello A, Sabatino R, et al. Interleukin-6 polymorphisms in HCC patients chronically infected with HCV. Infect Agent Cancer 2020;15:1-7.
https://doi.org/10.1186/s13027-020-00285-9 |
| 19 | Shah M, Danish L, Khan NU, Zaman F, Ismail M, Hussain M, et al. Determination of mutations in iron regulating genes of beta thalassemia major patients of Khyber Pakhtunkhwa, Pakistan. Mol Genet Genomic Med 2020;8:1-10.
https://doi.org/10.1002/mgg3.1310 |
| 20 | Border WA, Noble NA. Fibrosis linked to TGF-beta in yet another disease. J Clin Invest 1995;96:655-656.
https://doi.org/10.1172/JCI118107 |
| 21 | Kopp JB, Factor VM, Mozes M, Nagy P, Sanderson N, Böttinger EP, et al. Transgenic mice with increased plasma levels of TGF-β1 develop progressive renal disease. Lab Investig 1996;74:991-1003.
|
| 22 | Wong TYH, Poon P, Chow KM, Szeto CC, Cheung MK, Li PKT. Association of transforming growth factor-beta (TGF-β) T869C (Leu 10Pro) gene polymorphisms with type 2 diabetic nephropathy in Chinese. Kidney Int 2003;63:1831-1835.
https://doi.org/10.1046/j.1523-1755.2003.00919.x |
| 23 | Celedón JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, et al. The transforming growth factor-β1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 2004;13:1649-1656.
https://doi.org/10.1093/hmg/ddh171 |
| 24 | Buraczynska M, Baranowicz-Gaszczyk I, Borowicz E, Ksiazek A. TGF-β1 and TSC-22 gene polymorphisms and susceptibility to microvascular complications in type 2 diabetes. Nephron Physiol 2007;106:69-75.
https://doi.org/10.1159/000104874 |
| 25 | Akai Y, Sato H, Ozaki H, Iwano M, Dohi Y, Kanauchi M. Association of transforming growth factor-β1 T29C polymorphism with the progression of diabetic nephropathy. American Journal of Kidney Diseases. W.B. Saunders; 2001. DOI: 10.1053/ajkd.2001.27439
https://doi.org/10.1053/ajkd.2001.27439 |
| 26 | Babel N, Gabdrakhmanova L, Hammer MH, Schoenemann C, Skrypnikov V, Poliak N, et al. Predictive value of cytokine gene polymorphisms for the development of end-stage renal disease. J Nephrol 2006;19:802-807.
|
| 27 | Shah VN, Cheema BS, Sharma R, Khullar M, Kohli HS, Ahluwalia TS, et al. ACACβ gene (rs2268388) and AGTR1 gene (rs5186) polymorphism and the risk of nephropathy in Asian Indian patients with type 2 diabetes. Mol Cell Biochem 2013;372:191-198.
https://doi.org/10.1007/s11010-012-1460-2 |
| 28 | Muhammad A, Ullah N, Khan HU, Khan MJ. MicroRNAs in Lung Cancer tissues and Pleural fluids : A Review 2019;8:1-8.
|
| 29 | Moradi M, Rahimi Z, Amiri S, Rahimi Z, Vessal M, Nasri H. AT1R A1166C variants in patients with type 2 diabetes mellitus and diabetic nephropathy. J Nephropathol 2015;4:69-76.
|
| 30 | Mao S, Huang S. Association of angiotensinogen gene M235T polymorphism with the risk of IgA nephropathy: A meta-analysis. Ren Fail 2014;36:466-72.
https://doi.org/10.3109/0886022X.2013.868318 |