Single-Cell Analysis Reveals That CD47 mRNA Expression Correlates with Immune Cell Activation, Antiviral ISGs, and Cytotoxicity
bDepartment of Clinical Medicine, Aarhus University, Aarhus 8200, Denmark,
cDepartment of Biomedicine, Aarhus University, Aarhus 8000, Denmark
Keywords
Abstract
Background/Aims:
Immune cells are reported to upregulate CD47 during infection, however, the role of CD47 in innate and adaptive immune cells remains unclear.Methods:
To bridge this knowledge gap, we analysed our single cell (sc)-RNA dataset along with other publicly available sc-RNA datasets from healthy controls, people with HIV-1 (PWH) and COVID-19 patients. We characterized each immune cell based on low, intermediate, and high expression of CD47.Results:
Our analyses revealed that CD47high pDCs and monocytes exhibited relatively higher expression of IFN-α regulatory genes, antiviral interferon-stimulated genes (ISGs) and MHC-I associated genes compared to CD47inter. and CD47low cells. Furthermore, CD47high NK and CD8+ T cells showed higher expression of antiviral ISGs, as well as genes encoding for cytotoxic markers like granzyme B, perforin, granulysin, interferon gamma and NKG7. Additionally, CD47high CD8+ T cells expressed higher levels of PD-1 and LAG-3 genes. Lastly, we found that CD47high B cells had enriched expression of genes involved in cell activation and humoral responses.Conclusion:
Overall, our analyses revealed that innate and adaptive immune cells expressing elevated activation and functional gene signatures also express higher CD47 levels.Introduction
Integrin associated protein (CD47) is a widely and lowly expressed glycoprotein on the surface of all healthy cells [1-4]. CD47 is the ligand for signal regulatory protein alpha (SIRPα), an inhibitory receptor predominantly expressed in macrophages and myeloid dendritic cells. Acting as a self-marker to maintain homeostasis and ensure normal cell survival, the engagement of CD47 with SIRPα induces an anti-phagocytic “Don’t Eat-Me” signal [2, 5-7]. CD47/SIRPα binding results in the phosphorylation of immune-receptor tyrosine-based inhibitory motifs (ITIMs) within the cytoplasmic tail of SIRPα. Such phosphorylation leads to the recruitment and activation of inhibitory molecules such as Src homology 2 (SH2) domain- containing phosphatases (SHP-1 and SHP-2), which in turn regulate downstream signalling pathways, thus, resulting in inhibition of phagocytosis [8-11]. Another reported ligand of CD47 is thrombospondin-1 (TSP-1) [12]. The interaction between CD47 and TSP-1 reduces inflammation [13], limits primary CD8+ T cells antigen mediated activation and can cause differentiation of T cells into immunosuppressive phenotype [14].
Under normal physiological conditions, haematopoietic stem cells (HSCs) and progenitors are at constant risk of being phagocytosed by macrophages. To evade this attack, HSCs and progenitors transiently upregulate CD47 prior to and during migratory phases, and the level of CD47 determine the cells probability of survival [15, 16]. A similar immune evasion mechanism has also been observed in red blood cells (RBCs). Young RBCs overexpress CD47, while aged RBCs have reduced CD47 expression [17, 18] which may contribute to the clearance of aged RBCs.
CD47 has also become a hot research topic in oncology as it has been shown that tumor cells overexpress CD47 as a means to evade macrophage-mediated phagocytosis [19]. For instance, in ovarian cancer cells, haematological malignancies and other cancer types, elevated CD47 expression efficiently promotes tumor migration and tissue invasion. High CD47 expression is significantly correlated with poor prognosis in these cancer types [20-24]. In addition, observations in many different infections have shown that CD47 is upregulated in both immune cells and virus-infected cells, mainly due to Toll-like receptor (TLR) activation and induction by cytokines such as IFN-α, CXCL10, and TNF-α. Pro-inflammatory cytokines found in the plasma of hepatitis C patients were found to upregulate CD47 on uninfected dendritic cells. Importantly, CD47 upregulation on immune cells was not limited to virus infected cells but also occurred on surveilling immune cells in response to pathogen recognition [1, 25]. Additionally, viruses have evolved to exploit inhibitory signaling pathways to evade host immune surveillance [26]. Studies have reported that poxviruses encode expression of CD47-like protein which enhances their virulence by decreasing both macrophage and T cells activation [27, 28].
While significant progress has been made in elucidating the role of CD47 in immune regulation and disease pathogenesis, several challenges remain in the field. A comprehensive understanding of the dynamic regulation of CD47 expression and its functional consequences in diverse cell types and disease contexts is essential for the development of targeted therapeutic strategies aimed at modulating CD47 signaling pathways in cancer and infectious diseases. Moreover, the precise mechanisms underlying CD47 dysregulation during viral infections and its impact on immune cell function represent critical areas requiring further investigation. Our study aims to provide insights into the intricate interplay between CD47 expression levels and immune cell activation during viral infections, thereby advancing our understanding of CD47-mediated immune regulation and its potential implication for therapeutic interventions.
Materials and Methods
sc-RNA-seq data pre-processing and quality control
Publicly
available sc-RNA seq raw data were either downloaded or sent to us
upon request. Using Seurat packages (version 4.0), we performed a
clean-up and quality control based on cellular expression of
mitochondrial genes (cells with a percentage of mitochondrial genes
>5% were discarded). To be considered for further analysis, genes
had to be expressed in more than ten cells, cellular barcodes had to
be associated to at least 200 genes. We next identified doublets
using the DoubletFinder algorithm and removed these cell doublets
from the analysis. Each singlet file was saved for further analysis.
sc-RNA-seq data integration and identification of cell clusters
Next,
we integrated individual singlet dataset of healthy individuals,
HIV-1 infected individuals, COVID-19 patients or healthy with HIV-1
infected for the case of figure 1. In
detail, data was log1p-normalized with the SCT normalizeData method
using the “SCTransform” function, and subsequently scaled by the
Pearson Residuals with a scale factor of 10, 000 as default using the
“ScaleData” function. The top 2000 highly variable features were
selected using the “SelectIntegrationFeatures” function, followed
by finding the integration anchors using the “FindIntegrationAnchors”
function, performing the integration of the data using the
“IntegrateData” function. Following integration, principal
component analysis was performed using the “RunPCA” function with
default parameters, then both t-SNE (t-Distributed Stochastic
Neighbor Embedding) and UMAP (Uniform Manifold Approximation and
Projection) dimensionality reduction methods were conducted based on
the top 20 principal components (PCs) using the “RunTSNE” and
“RunUMAP” functions, respectively.
Next,
we normalized each integrated dataset, Variable
genes were determined using Seurat’s “FindVariableGenes”
function with default parameters (selection.method = “vst”,
nfeatures = 2000). Clusters were identified via the “FindClusters”
function (with
a cluster resolution of 0, 3)
implemented in Seurat using principal components with a P value
< 0.01. To
assign cellular identity, we applied graph-based clustering and a
non-linear dimension reduction using uniform manifold approximation
and projection (UMAP) or tSNE for cell cluster visualization.
using the “RunTSNE” and “RunUMAP” functions
(reduction = “pca”). Using
the differentially expressed genes for known lineage markers, we
annotated cell types based on these markers: CD8 T cells (CD3G,
CD3D, CD8A, IFIT2, GZMH), CD4 T cells (CD3G,
CD3D, CD4, IL7R, CCR7), monocytes
(LZY,
S100A8, S100A9, S100A12),
B cells (MS4A1,
CD79A, CD74),
Dendritic cells (IL3RA,
ITGAX, LST1, FCER1G, FCER3A),
platelet (PPBP),
NK cells, (GNLY,
NKG7),
pDC (CLEC4C,
LILRA4, IL3RA).
Division of cell type in to CD47low, CD47inter., and CD47high cells
All
immune cells, monocytes, pDC, NK cells, T cells and B cells were
categorized based on low, intermediate and high CD47
expression levels. This analysis was performed by first analyzing the
CD47
expression levels using volin plot and then set the criteria for the
different categories. For instance. Immune cells were categorized
into CD47low,
CD47inter.,
and CD47high
immune
cells using the “Subset” function. Example:
DefaultAssay(lamin_all)
<- "RNA"
lamin_all$CD47exp
<- count [,"CD47"]
lamin_all$CD47_level
<- "NA"
lamin_all$CD47_level
[which(lamin_all$CD47exp < 0.5)] <- "CD47 Low"
lamin_all$CD47_level
[which((lamin_all$CD47exp > 0.5) & (lamin_all$CD47exp <
1.5))] <- "CD47 Inter."
lamin_all$CD47_level
[which(lamin_all$CD47exp > 1.5)] <- "CD47 High"
table(lamin_all$CD47_level)
Idents(lamin_all)
<- "CD47_level"
levels(lamin_all)
levels(lamin_all)<-
c("CD47 Low", "CD47 Inter.", "CD47 High")
Each dataset of CD47low, CD47inter., and CD47high immune cells were saved for further analysis. Similar criteria were applied to divide monocytes, pDCs, NK cells, T cells and B cells.
Differentially expressed genes (DEGs)
“FindAllMarkers”
function implemented in Seurat v3 was used to identify DEGs across
clusters with the options “min.pct = 0.25, logfc.threshold = 0.25”.
Multiple test correction for P value
was performed using the Bonferroni method, and 0.05 was set as a
threshold to define significance. Furthermore, for analysis of
different CD47
expressing levels, average
gene expression was taken at the sample-level, followed by
differential gene expression analysis using DESeq2 r package across
the three groups (Wald test was applied for the analysis, followed by
the visualization using pheatmap R package.
Gene ontology enrichment analysis
Gene
Ontology (GO) analysis was performed using the clusterProfiler 4.0
package. The GO terms of selected genes were enriched in the database
“org.Hs.eg.db” using “enrichGO” function because of the lack
of study in pigs. Benjamini–Hochberg (BH) method was used for the
multiple test adjustment.
Quantification and Statistical Analysis
Differential
expression genes (DEGs) were analyzed using three tests,
Wilcoxon-ranked sum test, t-test and t-test overestimated variance.
DEGs were computed using the ‘FindMarker’ function of Seurat and
the probability values were estimated with respect to all other
clusters within each dataset.
Data availability
scRNA-seq
data that support the findings of this study are been deposited in
the Gene Expression Omnibus under (GEO
accession
number GSE157829),
(accession code GSE228078),
(GEO
accession
number GSE169346)
and (Array
Express with accession number: E-MTAB-9544).
Original
codes used to generate data of this paper are publicly available at
GitHub:
https://github.com/laminbcham/CD47-upregulation-on-human-immune-cells/blob/main/codes
Results
CD47high immune cells exhibit higher expression of immune activation and functional genes
To
investigate the role of CD47
in host immune cells, we used a publicly available sc-RNA dataset
(GEO accession
number GSE157829) [29].
This study reported single-cell RNA sequencing data from peripheral
blood mononuclear cells (PBMCs) from four healthy individuals (37,
847 cells) and six HIV-infected donors (28, 610 cells) [29].
We
processed the sc-RNA dataset and identified distinct immune cell
populations (Fig.
1A). To determine whether CD47
is upregulated at transcriptomic level during HIV-1 infection, we
analysed the CD47
expression in healthy individuals compared to PWH. We found that CD47
expression was upregulated during HIV-1 infection (Suppl. Fig. 1A).
Next, we investigated CD47
and SIRPα
expression and found that CD47
is expressed in all immune cells while its receptor (SIRPα) is
predominantly expressed in monocytes (Fig. 1B). Furthermore, our
analysis on CD47
expression levels in each immune cell type revealed that Natural
Killers (NK) cells, and CD8+ T cells have higher CD47
expression compared to other immune cell types (Fig. 1C).
To
determine the association between CD47
expression levels and immune cell function, we categorized all cells
based on their CD47
expression level: low (CD47<0.5),
intermediate (CD47
≥0.5
& CD47≤1.5)
and high (CD47>1.5)
(Fig. 1D). Detailed description is provided in the methods section.
Next, we determined the average gene expression profile of various
factors including IFN-α
regulatory genes (e.g. STAT1/2,
IRF7, IRF9, TLR7),
antiviral ISGs (e.g. IFI6,
MX1, ISG15, LYE6),
cytotoxic-related genes (e.g. IFNG,
GZMA, GZMB, NKG7,KLRB1, GNLY, PRF1)
and exhaustion-associated genes (e.g. PDCD1,
LAG-3, TIGIT)
among CD47high,
CD47inter.
and
CD47low
immune cells. Compared to CD47inter.
and
CD47low,
CD47high
immune cells exhibited higher expression of IFN-α
regulatory genes, antiviral ISGs, cellular activation, cytotoxic, and
exhaustion related genes (Figures 1E, F). Collectively, these
findings suggest that upregulation of CD47
was associated with immune activation, antiviral response,
cytotoxicity and exhaustion.
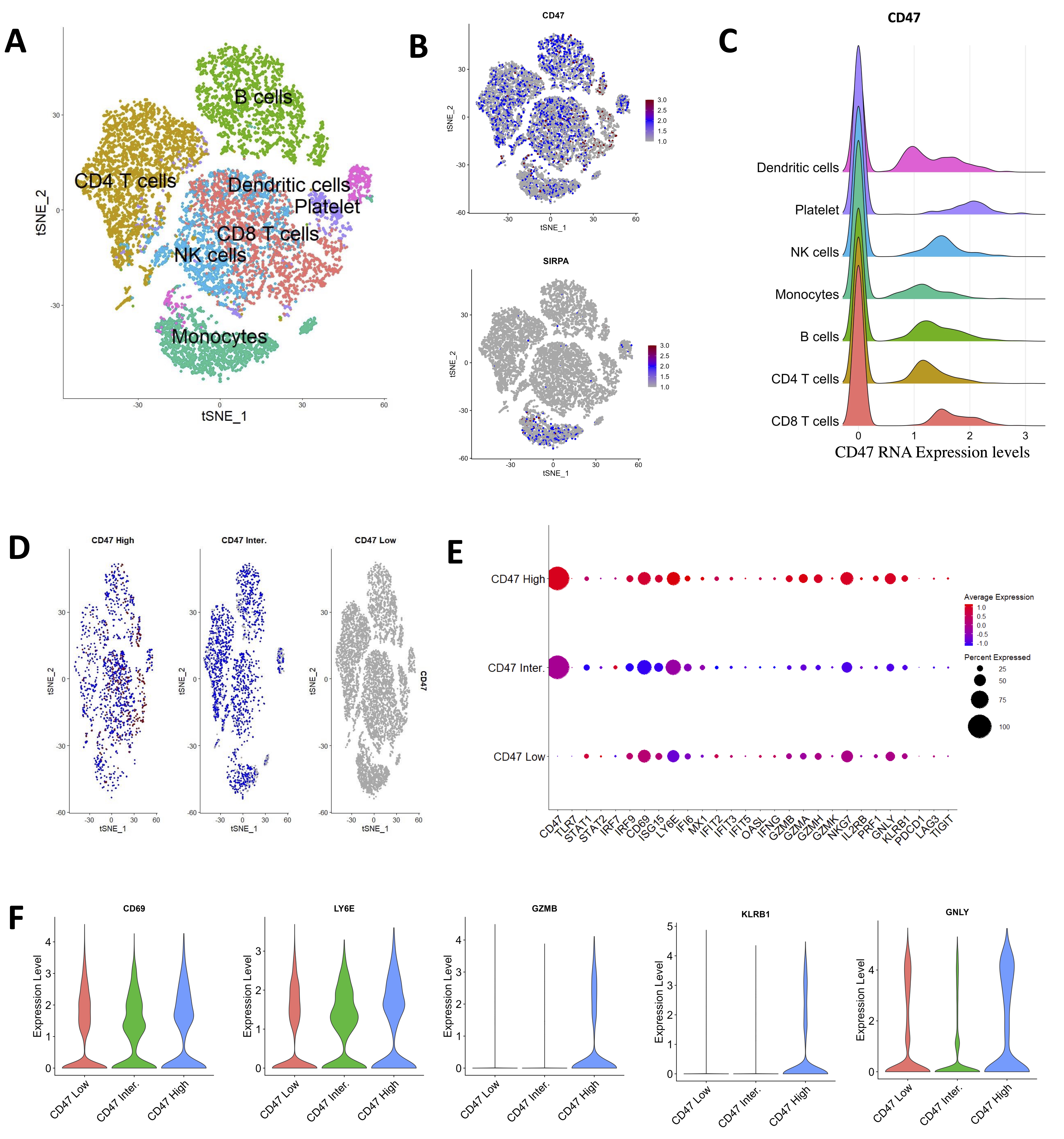
Fig. 1: CD47high immune cells exhibit higher expression of immune activation and functional genes. (A) tSNE representation of immune cells (n= 43,522 cells) from healthy and PWH. (B) tSNE and feature plot of CD47 and SIRPa expression levels on immune cells. (C) Ridge plot of CD47 expression in each immune cell type. Immune cells were divided into low, intermediate, and high CD47 expression levels (D) tSNE plot representation of CD47low, CD47 inter, and CD47high immune cells. (E) Dot plot representation of average and percentage expression of immune activation and functional genes and (F) Violin plot representation of CD69, LY6E, GZMB, KLRB1 and GNLY expression among CD47low, CD47inter., and CD47high immune cells.
CD47
upregulation on monocytes and pDCs is associated with higher
expression of IFN regulatory genes and antiviral ISGs
To
investigate the impact of CD47
expression levels on monocytes, we used a publicly available sc-RNA
dataset of
81, 643 antigen-presenting
cells
(APCs), including monocytes and dendritic cell (DC) subsets from
COVID-19 patients (GEO accession
number GSE169346)
[30].
After processing the sc-RNA dataset, our cluster analysis revealed
three main immune subsets: monocytes (CD14 and CD16 monocytes),
conventional dendritic cells (cDC) (CD1c, CLEC9A and AS dendritic
cells) and pDCs (Suppl. Fig. 1B). Among
these APC subsets, CD14+ monocytes were found to express lower CD47
levels compared to all other APC subsets
(Suppl. Fig. 1C). To examine how CD47
expression levels influence monocyte activation and function, we
categorized monocytes based on low, intermediate, and high CD47
expression levels and analysed the hallmark gene set. We found that
CD47high
monocytes had relatively higher expression of genes involved in the
IFN-α
response, complement response, inflammatory response, IFN-γ
response, IL2_STAT5 response, and TNF-α
response (Fig. 2A). Further, a differential gene expression and gene
ontology analysis between CD47high
and CD47low
monocytes showed enrichment of genes involved in type I interferon
production, cellular response to interferon and chemokine-mediated
signalling pathways in CD47high
compared to CD47low
monocytes (Suppl. Fig. 1D, E). To further confirm the association
between CD47
upregulation and monocyte activation and function, we categorized
monocytes based on low and high expression of IRF7,
IFNAR1, MX1, ISG15, CD86, IL1B, NKG7
and TNF
genes
and
then analysed the CD47
expression levels. We found that monocytes expressing high levels of
these genes also upregulated their CD47
expression (Fig. 2B), thus, indicating that highly activated and
functional monocytes have elevated CD47
expression levels.
Next,
we investigated the role of CD47
on pDCs using our sc-RNA dataset of enriched pDCs from PWH (GEO
accession
number GSE228078)
[31].
After reprocessing the dataset, we performed cluster analysis (Suppl.
Fig. 2A) and re-cluster the pDC subsets (Suppl. Fig. 2B). We found
that pDC1 and cytotoxic-pDC cluster had higher expression of CD47
(Suppl. Fig. 2C). Next, we categorized pDCs based on low,
intermediate, and high CD47
expression levels and evaluated the activation and functional gene
signatures. We found that 69, 1% were classified as CD47low,
18.3% as CD47inter.
and 12.6% as
CD47high
pDCs (Suppl. Fig. 2D). Our data showed that CD47high
and CD47inter.
pDCs displayed higher expression of IFN-α
regulatory genes compared to CD47low
pDCs. However, we observed higher expression of antiviral ISGs, and
MHC-I associated genes in CD47high
pDCs compared to CD47inter
and CD47low
pDCs (Fig. 2C, D). Gene ontology analysis showed that CD47high
pDCs were enriched with gene transcripts involved in IFN production,
cellular response to IFN and defense against virus (Fig. 2E).
Overall, our results highlight that the upregulation of CD47
in monocytes and pDCs is associated with enhanced innate immune
activation and function.
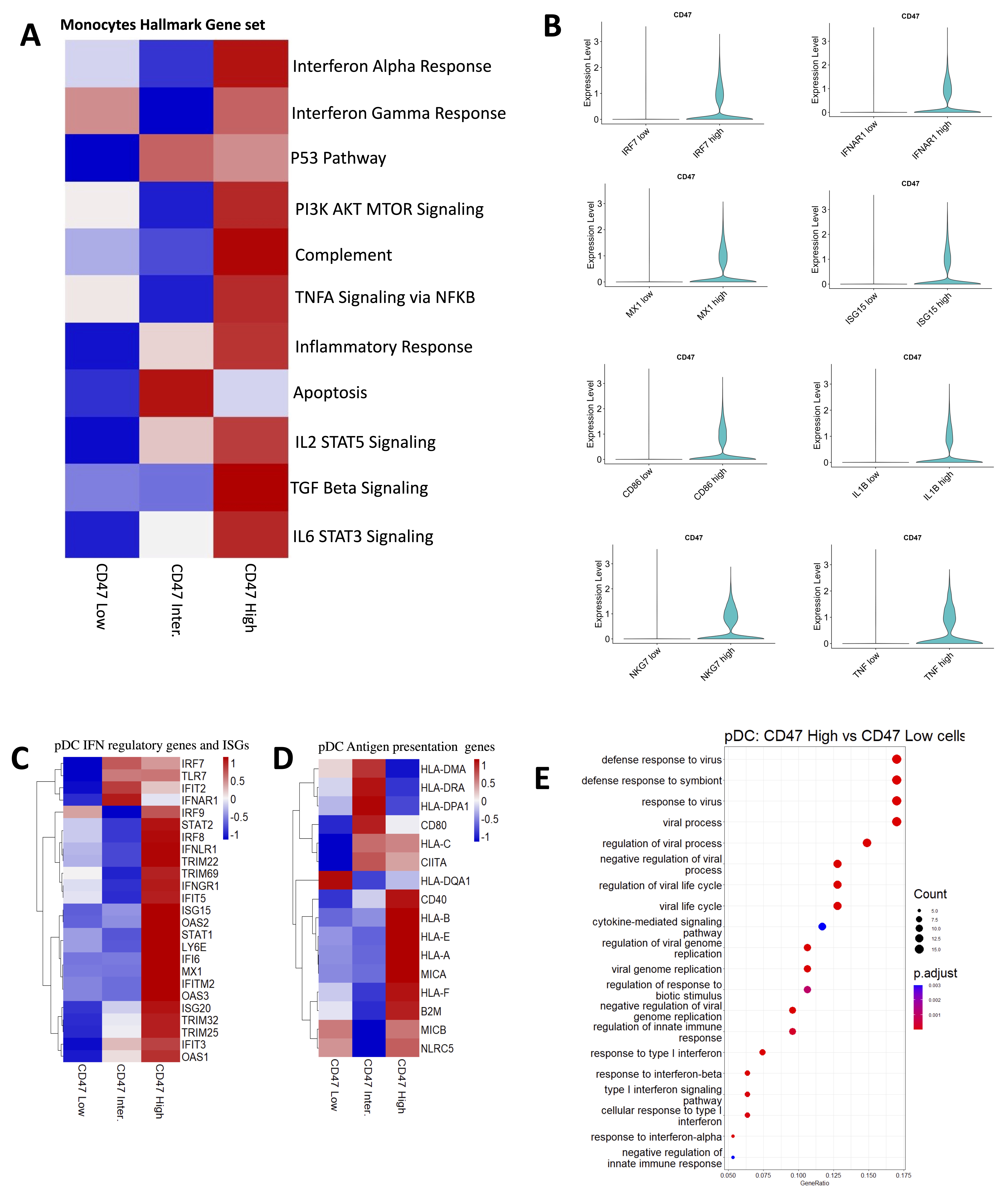
Fig. 2: CD47 upregulation on monocytes and pDCs is associated with higher expression of IFN regulatory genes and antiviral ISGs. Monocytes were divided into low, intermediate and high CD47 expression levels. (A) Heatmap illustration of relative gene expression levels of Hallmark gene set among CD47low, CD47inter., and CD47high monocytes. (B) Violin plot representation of CD47 expression levels among monocytes expressing low or high IRF7, IFNAR1, MX1, ISG15, CD86, IL1B, NKG7 and TNF. Heatmap illustration of relative gene expression levels of (C) pDCs IFN regulatory genes and ISGs and (D) pDCs antigen presentation genes among CD47low, CD47inter., and CD47high pDCs. (E) Dot plot representation of gene ontology between CD47low and CD47high pDCs.
CD47high
NK cells have higher expression of ISGs and cytotoxic encoding-genes
To
determine the impact of CD47
expression levels on NK cell activation and function, we interrogated
NK cells from our sc-RNA dataset (Suppl. Fig. 2A) (GEO accession
number GSE228078)[31].
The cluster analysis revealed four NK clusters: CD56dim,
CD56bright,
NKT and CD16negative
NK cells (Fig. 3A). We observed that CD56dim,
CD16neg
and NK/T cells have higher CD47
expression levels compared to CD56bright
NK cells (Fig. 3B). To establish the association between CD47
expression and NK cells activation and function, we categorized NK
cells into low, intermediate and high CD47
expression levels. Our analysis of antiviral and cytotoxic genes
expression showed that CD47high
NK cells have relatively higher expression of KLRD1,
FCGR3A, MX1, ISG15, IFI6, NKG7, GNLY, PRF1, GZMB
(Fig. 3C). To further validate these findings, we categorized NK
cells into low or high expression of MX1,
NKG7, PRF1, GZMB
and GNLY,
and analysed their CD47
expression levels. Our data revealed that NK cells expressing high
levels of NKG7
PRF1, GZMB
and GNLY
also expressed higher levels of CD47
(Fig. 3D). Together, our findings indicate that highly activated and
cytotoxic NK cells also have elevated CD47
expression.
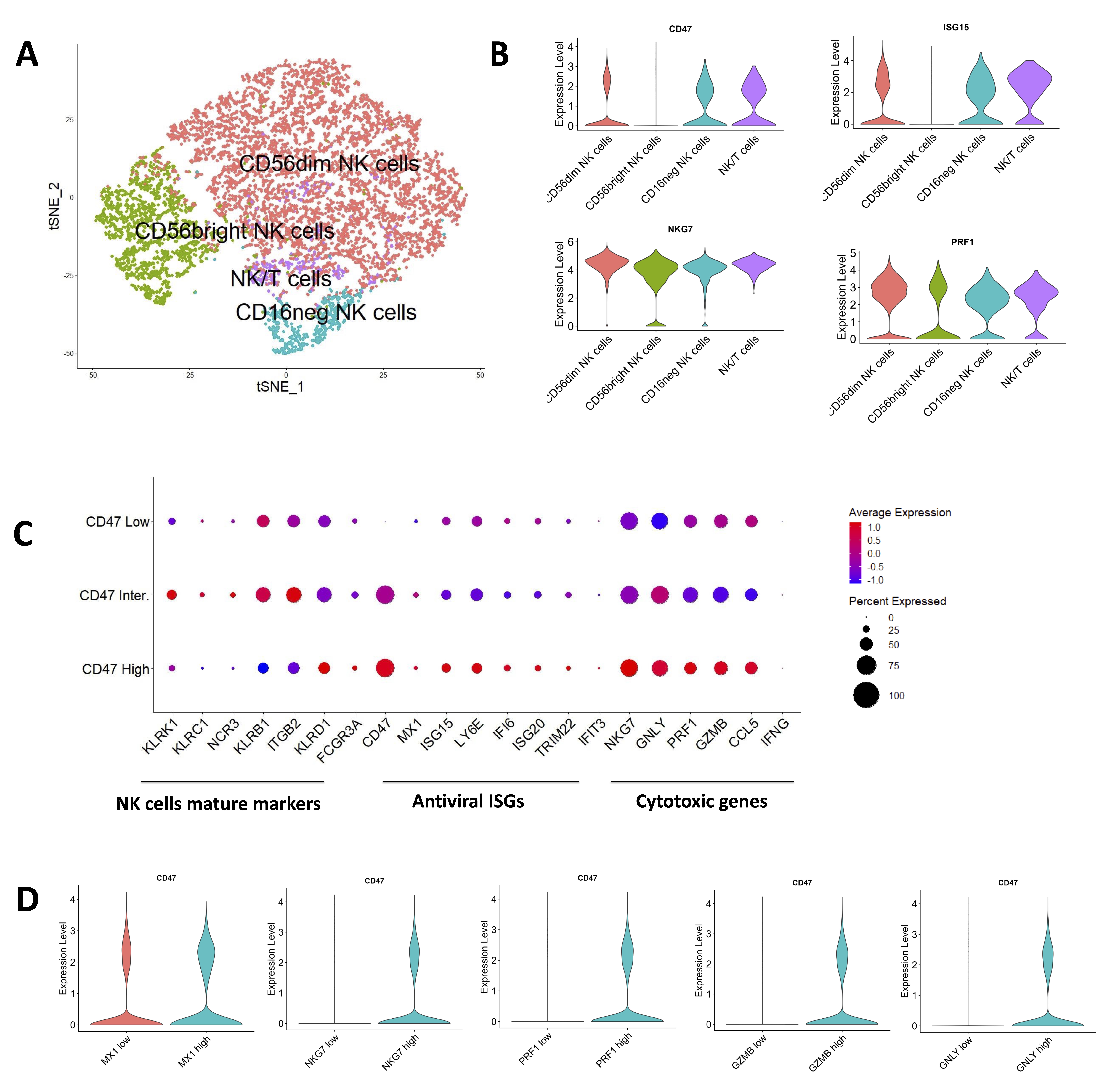
Fig. 3: CD47high NK cells have higher expression of ISGs and cytotoxic encoding-genes. (A) tSNE illustration of NK cells subsets (n= 18,032 NK cells) from PWH. (B) Violin plot representation of CD47, ISG15, NKG7 and PRF1 expression among NK cells subsets. NK cells were divided into low, intermediate and high CD47 expression levels. (C) Dot plot representation of average and percentage expression of NK cell mature markers, antiviral ISGs and cytotoxic genes among NK CD47low, CD47inter., and CD47high. (F) Violin plot representation of CD47 expression levels among NK cells expressing low or high expression of MX1, NKG7, PRF1, GZMB, and GNLY genes.
CD47
upregulation on T cells is associated with increased activation,
cytotoxicity and exhaustion
Similar
to NK cells, we examined the impact of CD47
expression on T cells. First, we selected the CD4+ and CD8+ T cells
from our sc-RNA dataset from PWH (Suppl. Fig. 2A) (GEO accession
number GSE228078)[31].
The cluster analysis identified CD4+ and CD8+ T cells subsets (Suppl.
Fig. 3A and Fig. 4A). Our analysis showed that CD47
expression was higher in cytotoxic CD4+ T cells compared to other
CD4+ T cells subsets (Suppl. Fig. 3B). Similarly, effector CD8+ T
cells showed elevated CD47
and GZMA
expression levels in comparison to other CD8+ T cells subsets (Fig.
4B). To determine whether the antiviral and cytotoxicity capabilities
of CD8+ T cells is associated with CD47
upregulation, we categorized CD8+ T cells into low, intermediate, and
high CD47
expression levels and analysed activation, cytotoxic and exhaustion
encoding genes. Our data revealed that CD47high
CD8+ T cells have relatively higher expression of CD69,
IFI6, ISG15, ISG20, TNFSF14, GZMB, NKG7, IL2RB, IFNG, PDCD1, LAG-3
etc., while CD47inter.
CD8+ T cells exhibit higher expression of CD38,
OAS1
and TNFRSF9
(Fig. 4C, D). To further confirm this association, we divided CD8+ T
cells into low or high ISG15
and PRF1
expression and analysed CD47
expression levels. We found that CD8+ T cells expressing high levels
of ISG15
or PRF1
also have elevated CD47
expression levels (Fig. 4E). Overall, our data showed that
upregulation of CD47
in T cells is associated with T cells activation, cytotoxicity, and
exhaustion.
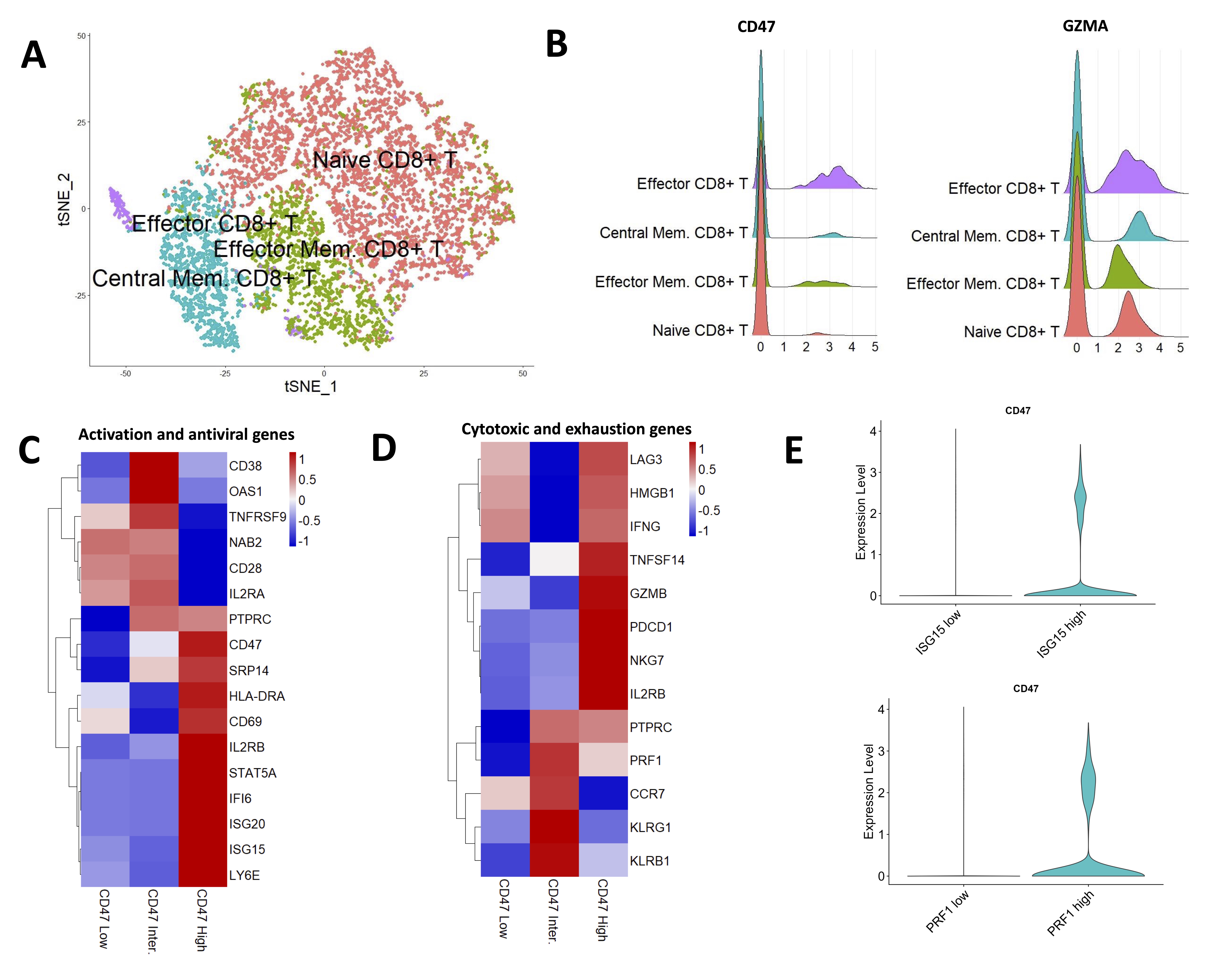
Fig. 4: CD47 upregulation on T cells is associated with increased activation, cytotoxicity and exhaustion. (A) tSNE plot illustration of CD8+ T cells subsets (n= 11,271 CD8+ T cells) from PWH. (B) Ridge plot of CD47 and GZMB expression among CD8+ T cells subsets. CD8+ T cells were divided into low, intermediate and high CD47 expression levels. Heatmap illustration of relative gene expression levels of CD8+ T cells (C) activation genes and ISGs and (D) cytotoxic and exhaustion genes among CD47low, CD47inter., and CD47high CD8+ T cells. (F) Violin plot representation of CD47 expression levels among CD8+ T cells expressing low or high expression of ISG15, and PRF1 genes.
B
cells with elevated CD47 are enriched with genes involved in B cells
activation and humoral response
To
understand how CD47
expression levels impact B cell activation and response, we used a
publicly available sc-RNA dataset
based on phenotypically sorted B cells
(Array
Express with accession number: E-MTAB-9544)
[32].
This sc-RNA dataset enabled us to perform an in-depth
characterization of a large number of B cells. After processing the
sc-RNA dataset and performing cluster analysis, we identified five B
cells subsets: naïve, transitional, double negative, classical
memory and IgM memory B cells subsets (Fig. 5A). CD47
expression was similar in all B cell subsets (Fig. 5B). Next, we
categorized B cells based on low, intermediate, and high CD47
expression levels and analysed B cells activation and functional
encoding genes. These analyses showed that CD47high
B cells have relatively higher expression of TLR9,
IRF7, MZB1, ISG15, MX1, OAS1, CD40, CD80, HLA-C, IGHM,
while CD47inter
B cells displayed higher expression of STAT1/2,
IGHD, HLA-B, HLA-DMA
and HLA-DR
compared to CD47low
B cells (Fig. 5C). Differential gene expression and gene ontology
analysis showed that CD47high
B cells were enriched for gene transcripts involved in the regulation
of B cells activation, humoral response, immunoglobulin-mediated
response, response to chemokine, cellular response to IFN-α
and IFN-γ,
antigen-mediated signalling, and programmed cell death (Fig. 5D).
Overall, our data highlighted that the upregulation of CD47
on B cells is associated with increased expression of activation and
antiviral genes as well as genes involved in mediating humoral B cell
response.
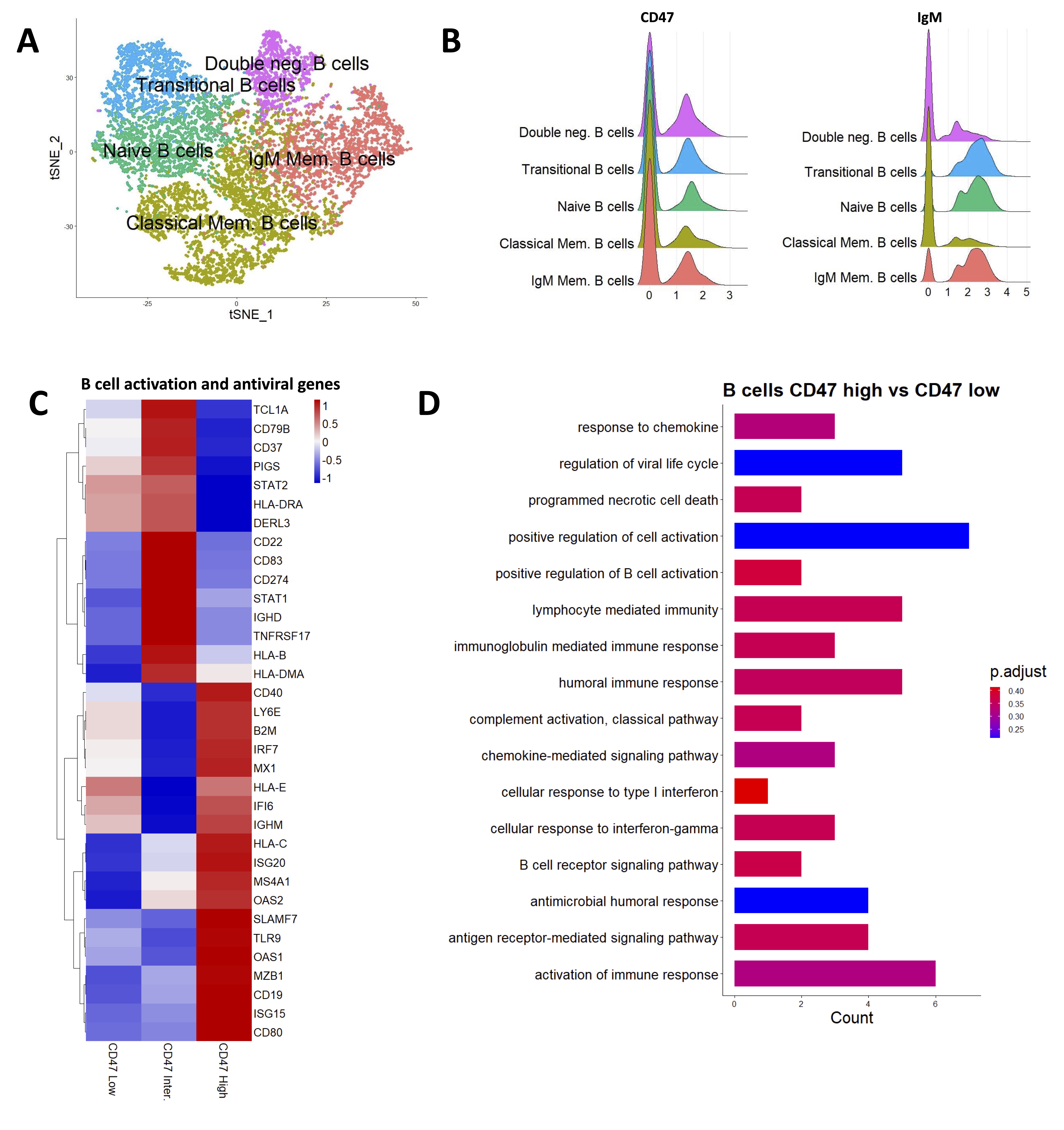
Fig. 5: B cells with elevated CD47 are enriched with genes involved in B cells activation and humoral response: (A) tSNE plot illustration of B cells subsets (n= 41,085 B cells) from healthy individuals. (B) Ridge plot of CD47 and IgM expression among B cells subsets. B cells were divided into low, intermediate and high CD47 expression levels. (C) Heatmap illustration of relative gene expression levels of B cells activation genes, ISGs and functional genes among CD47low, CD47inter., and CD47high B cells. (D) Bar plot representation of gene ontology between CD47low and CD47high B cells.
Discussion
Previous studies have demonstrated that immune cells and virus-infected cells upregulate CD47 during infection [25, 33]. In light of this, we investigated the association between CD47 upregulation and immune cell activation and function using sc-RNA dataset from healthy controls, PWH and COVID-19 patients. Our comprehensive analysis revealed that CD47 upregulation in monocytes and pDCs is associated with relatively higher expression of genes involved in IFN-α production, cellular response to IFN-α, as well as increased antigen presentation. Similarly, NK cell and T cells expressing high levels of CD47 also exhibit high expression of genes encoding for cellular activation, antiviral ISGs, cytotoxicity, and exhaustion. Additionally, the upregulation of CD47 on B cells is also linked to enhanced B cells activation and humoral response.
CD47 has previously been defined as an interferon-stimulated gene, and its upregulation is part of a coordinated program of host defence mechanisms triggered by IFN-α production [25, 34-36]. Our findings align with these studies, demonstrating that the upregulation of CD47 in monocytes, pDCs, NK cells, T cells and B cells is associated with relatively higher expression of antiviral ISGs. Moreover, our study findings shed new light that upregulation of CD47 on host immune cells is an indicating marker of activation, function and possibly exhaustion. Our data reveals that CD47 upregulation is not just limited to IFN-α mediated ISGs induction but also highlights the upregulation of CD47 in IFN-α producing cells. Monocytes and pDCs with intermediate or high expression of CD47 have relatively higher expression of genes involve in type I interferon production, chemokine mediated signaling, complement activation and cellular response to other cytokines such as IFN-α and TNF-α. These findings suggest transient upregulation of CD47 upon TLR activation, consistent with previous report [25]. Similarly, NK cell and CD8+ T cells with intermediate or high CD47 levels also express higher levels of GZMB, IFNG, PRF1 and NKG7, clearly indicating that highly cytotoxic NK cells and T cells upregulate CD47. Further aligning with reports that CD47 upregulation can also be induce by IFN-α and other unknown mechanisms [25]. Although PD-1 expression is linked to both T cell exhaustion as well activation [37, 38], our study reveals that CD47high CD8+ T cells express higher levels of PDCD1 and LAG-3 genes. In line with its effects on other immune cells, CD47 upregulation on B cells is linked with enhanced B cells activation and humoral immune response. Therefore, suggesting a direct correlation between CD47 expression levels and immune activation and function.
The question remains: why is CD47 upregulated in highly activated and functional immune cells? Given the well-known anti-phagocytic protective function of CD47, it is reasonable to speculate that highly activated and functional immune cells increase CD47 expression as a defensive mechanism against phagocytosis. A recent study reported that CD47 transcriptional interference by HIV-1 Viral protein U (Vpu) might promote the susceptibility of macrophages to viral infection via phagocytosis of infected CD4+ T cells. The study highlighted that Vpu downregulates CD47 expression on infected CD4+ T cells, leading to enhanced capture and phagocytosis by macrophages [39]. These findings support our hypothesis that activated and functional host immune cells upregulate CD47 for protection. In addition, it has been reported that CD47-negative CAR-T cells fail to expand and persist in vivo due to continuous macrophage mediated phagocytosis, underscoring the importance of CD47 expression for CAR-T cell survival [40-42]. However, additional experiments are needed to elucidate how CD47 expression on each immune cell protects them from macrophage mediated phagocytosis.
Conclusion
In summary, our transcriptomic analysis of immune cells suggests a potential association between CD47 upregulation and cellular activation, antiviral ISGs, and cytotoxicity. Modulating CD47 expression levels may offer a novel approach for host-directed therapies, such as downregulating CD47 in HIV-1 infected CD4+ T cells as a part of the ‘’shock and kill’’ strategy or increasing CD47 expression on CAR-T cells therapy.
Acknowledgements
This research was funded by The Lundbeck Foundation, grant R313-2019-790. The authors wish to thank all research groups that allow us to reuse their raw sc-RNA data. We also wish to thank NGS Core Center, Department of Molecular Medicine, Aarhus University Hospital, Denmark for performing the next-generation sequencing.
Author Contributions
Conceptualization:
L.B.C, and., O.S.S.; methodology, L.B.C., M.R.U., L.L., M.T. and
O.S.S.; investigation, L.B.C., M.R.U., L.L., M.T. and O.S.S.;
verification, L.B.C., L.L., M.T. and O.S.S.; formal analysis, L.B.C.,
L.L., and O.S.S.; writing—original draft, L.B.C., and O.S.S.;
writing—review and editing, L.B.C., M.R.U., L.L., M.T. and O.S.S.;
visualization, L.B.C. and O.S.S.; resources, O.S.S.; supervision,
L.L. and O.S.S.; project administration, L.B.C. and O.S.S.; funding
acquisition, O.S.S. All authors have read and agreed to the published
version of the manuscript.
Disclosure Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interest exists.
References
| 1 | Cham LB, Adomati T, Li F, Ali M, Lang KS: CD47 as a Potential Target to Therapy for Infectious Diseases. Antibodies (Basel) 2020;9.
https://doi.org/10.3390/antib9030044 |
| 2 | Catalan R, Orozco-Morales M, Hernandez-Pedro NY, Guijosa A, Colin-Gonzalez AL, Avila-Moreno F, Arrieta O: CD47-SIRPalpha Axis as a Biomarker and Therapeutic Target in Cancer: Current Perspectives and Future Challenges in Nonsmall Cell Lung Cancer. J Immunol Res 2020;2020:9435030.
https://doi.org/10.1155/2020/9435030 |
| 3 | Lv Z, Bian Z, Shi L, Niu S, Ha B, Tremblay A, Li L, Zhang X, Paluszynski J, Liu M, Zen K, Liu Y: Loss of Cell Surface CD47 Clustering Formation and Binding Avidity to SIRPalpha Facilitate Apoptotic Cell Clearance by Macrophages. J Immunol 2015;195:661-671.
https://doi.org/10.4049/jimmunol.1401719 |
| 4 | Oldenborg PA: CD47: A Cell Surface Glycoprotein Which Regulates Multiple Functions of Hematopoietic Cells in Health and Disease. ISRN Hematol 2013;2013:614619.
https://doi.org/10.1155/2013/614619 |
| 5 | Toledano N, Gur-Wahnon D, Ben-Yehuda A, Rachmilewitz J: Novel CD47: SIRPalpha dependent mechanism for the activation of STAT3 in antigen-presenting cell. PLoS One 2013;8:e75595.
https://doi.org/10.1371/journal.pone.0075595 |
| 6 | Li F, Lv B, Liu Y, Hua T, Han J, Sun C, Xu L, Zhang Z, Feng Z, Cai Y, Zou Y, Ke Y, Jiang X: Blocking the CD47-SIRPalpha axis by delivery of anti-CD47 antibody induces antitumor effects in glioma and glioma stem cells. Oncoimmunology 2018;7:e1391973.
https://doi.org/10.1080/2162402X.2017.1391973 |
| 7 | Li C, Liu Y, Li D, Wang Q, Zhou S, Zhang H, Wang Y, He Z, Liu H, Sun J: Promising alternatives of CD47 monoclonal antibody: an injectable degradable hydrogel loaded with PQ912 for postoperative immunotherapy effectively blocks CD47-SIRPalpha signal. Theranostics 2022;12:4581-4598.
https://doi.org/10.7150/thno.72310 |
| 8 | Morrissey MA, Kern N, Vale RD: CD47 Ligation Repositions the Inhibitory Receptor SIRPA to Suppress Integrin Activation and Phagocytosis. Immunity 2020;53:290-302 e296.
https://doi.org/10.1016/j.immuni.2020.07.008 |
| 9 | Oldenborg PA, Gresham HD, Lindberg FP: CD47-signal regulatory protein alpha (SIRPalpha) regulates Fcgamma and complement receptor-mediated phagocytosis. J Exp Med 2001;193:855-862.
https://doi.org/10.1084/jem.193.7.855 |
| 10 | Qu T, Li B, Wang Y: Targeting CD47/SIRPalpha as a therapeutic strategy, where we are and where we are headed. Biomark Res 2022;10:20.
https://doi.org/10.1186/s40364-022-00373-5 |
| 11 | Bian Z, Shi L, Guo YL, Lv Z, Tang C, Niu S, Tremblay A, Venkataramani M, Culpepper C, Li L, Zhou Z, Mansour A, Zhang Y, Gewirtz A, Kidder K, Zen K, Liu Y: Cd47-Sirpalpha interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc Natl Acad Sci U S A 2016;113:E5434-5443.
https://doi.org/10.1073/pnas.1521069113 |
| 12 | Koduru SV, Sun BH, Walker JM, Zhu M, Simpson C, Dhodapkar M, Insogna KL: The contribution of cross-talk between the cell-surface proteins CD36 and CD47-TSP-1 in osteoclast formation and function. J Biol Chem 2018;293:15055-15069.
https://doi.org/10.1074/jbc.RA117.000633 |
| 13 | Lamy L, Foussat A, Brown EJ, Bornstein P, Ticchioni M, Bernard A: Interactions between CD47 and thrombospondin reduce inflammation. J Immunol 2007;178:5930-5939.
https://doi.org/10.4049/jimmunol.178.9.5930 |
| 14 | Stirling ER, Terabe M, Wilson AS, Kooshki M, Yamaleyeva LM, Alexander-Miller MA, Zhang W, Miller LD, Triozzi PL, Soto-Pantoja DR: Targeting the CD47/thrombospondin-1 signaling axis regulates immune cell bioenergetics in the tumor microenvironment to potentiate antitumor immune response. J Immunother Cancer 2022;10.
https://doi.org/10.1136/jitc-2022-004712 |
| 15 | Nonino A, Nascimento JM, Mascarenhas CC, Mazzeu JF, Pereira RW, Jacomo RH: CD47 expression is decreased in hematopoietic progenitor cells in patients with myelofibrosis. Braz J Med Biol Res 2018;52:e7784.
https://doi.org/10.1590/1414-431x20187784 |
| 16 | Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL: CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 2009;138:271-285.
https://doi.org/10.1016/j.cell.2009.05.046 |
| 17 | Burger P, Hilarius-Stokman P, de Korte D, van den Berg TK, van Bruggen R: CD47 functions as a molecular switch for erythrocyte phagocytosis. Blood 2012;119:5512-5521.
https://doi.org/10.1182/blood-2011-10-386805 |
| 18 | Torrez Dulgeroff LB, Oakley MS, Tal MC, Yiu YY, He JQ, Shoham M, Majam V, Okoth WA, Malla P, Kumar S, Weissman IL: CD47 blockade reduces the pathologic features of experimental cerebral malaria and promotes survival of hosts with Plasmodium infection. Proc Natl Acad Sci U S A 2021;118
https://doi.org/10.1073/pnas.1907653118 |
| 19 | Huang CY, Ye ZH, Huang MY, Lu JJ: Regulation of CD47 expression in cancer cells. Transl Oncol 2020;13:100862.
https://doi.org/10.1016/j.tranon.2020.100862 |
| 20 | Li Y, Lu S, Xu Y, Qiu C, Jin C, Wang Y, Liu Z, Kong B: Overexpression of CD47 predicts poor prognosis and promotes cancer cell invasion in high-grade serous ovarian carcinoma. Am J Transl Res 2017;9:2901-2910.
|
| 21 | Wang CL, Lin MJ, Hsu CY, Lin HY, Tsai HP, Long CY, Tsai EM, Hsieh TH, Wu CH: CD47 promotes cell growth and motility in epithelial ovarian cancer. Biomed Pharmacother 2019;119:109105.
https://doi.org/10.1016/j.biopha.2019.109105 |
| 22 | Yu L, Ding Y, Wan T, Deng T, Huang H, Liu J: Significance of CD47 and Its Association With Tumor Immune Microenvironment Heterogeneity in Ovarian Cancer. Front Immunol 2021;12:768115.
https://doi.org/10.3389/fimmu.2021.768115 |
| 23 | Eladl E, Tremblay-LeMay R, Rastgoo N, Musani R, Chen W, Liu A, Chang H: Role of CD47 in Hematological Malignancies. J Hematol Oncol 2020;13:96.
https://doi.org/10.1186/s13045-020-00930-1 |
| 24 | Sun J, Chen Y, Lubben B, Adebayo O, Muz B, Azab AK: CD47-targeting antibodies as a novel therapeutic strategy in hematologic malignancies. Leuk Res Rep 2021;16:100268.
https://doi.org/10.1016/j.lrr.2021.100268 |
| 25 | Tal MC, Torrez Dulgeroff LB, Myers L, Cham LB, Mayer-Barber KD, Bohrer AC, Castro E, Yiu YY, Lopez Angel C, Pham E, Carmody AB, Messer RJ, Gars E, Kortmann J, Markovic M, Hasenkrug M, Peterson KE, Winkler CW, Woods TA, Hansen P, et al.: Upregulation of CD47 Is a Host Checkpoint Response to Pathogen Recognition. mBio 2020;11
https://doi.org/10.1128/mBio.01293-20 |
| 26 | Farre D, Martinez-Vicente P, Engel P, Angulo A: Immunoglobulin superfamily members encoded by viruses and their multiple roles in immune evasion. Eur J Immunol 2017;47:780-796.
https://doi.org/10.1002/eji.201746984 |
| 27 | Cameron CM, Barrett JW, Mann M, Lucas A, McFadden G: Myxoma virus M128L is expressed as a cell surface CD47-like virulence factor that contributes to the downregulation of macrophage activation in vivo. Virology 2005;337:55-67.
https://doi.org/10.1016/j.virol.2005.03.037 |
| 28 | Cameron CM, Barrett JW, Liu L, Lucas AR, McFadden G: Myxoma virus M141R expresses a viral CD200 (vOX-2) that is responsible for down-regulation of macrophage and T-cell activation in vivo. J Virol 2005;79:6052-6067.
https://doi.org/10.1128/JVI.79.10.6052-6067.2005 |
| 29 | Wang S, Zhang Q, Hui H, Agrawal K, Karris MAY, Rana TM: An atlas of immune cell exhaustion in HIV-infected individuals revealed by single-cell transcriptomics. Emerg Microbes Infect 2020;9:2333-2347.
https://doi.org/10.1080/22221751.2020.1826361 |
| 30 | Saichi M, Ladjemi MZ, Korniotis S, Rousseau C, Ait Hamou Z, Massenet-Regad L, Amblard E, Noel F, Marie Y, Bouteiller D, Medvedovic J, Pene F, Soumelis V: Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat Cell Biol 2021;23:538-551.
https://doi.org/10.1038/s41556-021-00681-2 |
| 31 | Cham LB, Gunst JD, Schleimann MH, Frattari GS, Rosas-Umbert M, Vibholm LK, van der Sluis RM, Jakobsen MR, Olesen R, Lin L, Tolstrup M, Sogaard OS: Single cell analysis reveals a subset of cytotoxic-like plasmacytoid dendritic cells in people with HIV-1. iScience 2023;26:107628.
https://doi.org/10.1016/j.isci.2023.107628 |
| 32 | Stewart A, Ng JC, Wallis G, Tsioligka V, Fraternali F, Dunn-Walters DK: Single-Cell Transcriptomic Analyses Define Distinct Peripheral B Cell Subsets and Discrete Development Pathways. Front Immunol 2021;12:602539.
https://doi.org/10.3389/fimmu.2021.602539 |
| 33 | Cham LB, Torrez Dulgeroff LB, Tal MC, Adomati T, Li F, Bhat H, Huang A, Lang PA, Moreno ME, Rivera JM, Galkina SA, Kosikova G, Stoddart CA, McCune JM, Myers LM, Weissman IL, Lang KS, Hasenkrug KJ: Immunotherapeutic Blockade of CD47 Inhibitory Signaling Enhances Innate and Adaptive Immune Responses to Viral Infection. Cell Rep 2020;31:107494.
https://doi.org/10.1016/j.celrep.2020.03.058 |
| 34 | Ye ZH, Jiang XM, Huang MY, Xu YL, Chen YC, Yuan LW, Huang CY, Yu WB, Chen X, Lu JJ: Regulation of CD47 expression by interferon-gamma in cancer cells. Transl Oncol 2021;14:101162.
https://doi.org/10.1016/j.tranon.2021.101162 |
| 35 | Wang S, Xie L, Xie Z, Wan L, Huang J, Deng X, Xie ZQ, Luo S, Zeng T, Zhang Y, Zhang M, Zhou L: Dynamic Changes in the Expression of Interferon-Stimulated Genes in Joints of SPF Chickens Infected With Avian Reovirus. Front Vet Sci 2021;8:618124.
https://doi.org/10.3389/fvets.2021.618124 |
| 36 | Qu S, Jiao Z, Lu G, Xu J, Yao B, Wang T, Wang J, Yao Y, Yan X, Wang T, Liang H, Zen K: Human lung adenocarcinoma CD47 is upregulated by interferon-gamma and promotes tumor metastasis. Mol Ther Oncolytics 2022;25:276-287.
https://doi.org/10.1016/j.omto.2022.04.011 |
| 37 | Simon S, Labarriere N: PD-1 expression on tumor-specific T cells: Friend or foe for immunotherapy? Oncoimmunology 2017;7:e1364828.
https://doi.org/10.1080/2162402X.2017.1364828 |
| 38 | Jubel JM, Barbati ZR, Burger C, Wirtz DC, Schildberg FA: The Role of PD-1 in Acute and Chronic Infection. Front Immunol 2020;11:487.
https://doi.org/10.3389/fimmu.2020.00487 |
| 39 | Cong L, Sugden SM, Leclair P, Lim CJ, Pham TNQ, Cohen EA: HIV-1 Vpu Promotes Phagocytosis of Infected CD4(+) T Cells by Macrophages through Downregulation of CD47. mBio 2021;12:e0192021.
https://doi.org/10.1128/mBio.01920-21 |
| 40 | Beckett AN, Chockley P, Pruett-Miller SM, Nguyen P, Vogel P, Sheppard H, Krenciute G, Gottschalk S, DeRenzo C: CD47 expression is critical for CAR T-cell survival in vivo. J Immunother Cancer 2023;11.
https://doi.org/10.1136/jitc-2022-005857 |
| 41 | Chen H, Yang Y, Deng Y, Wei F, Zhao Q, Liu Y, Liu Z, Yu B, Huang Z: Delivery of CD47 blocker SIRPalpha-Fc by CAR-T cells enhances antitumor efficacy. J Immunother Cancer 2022;10.
https://doi.org/10.1136/jitc-2021-003737 |
| 42 | Dacek MM, Kurtz KG, Wallisch P, Pierre SA, Khayat S, Bourne CM, Gardner TJ, Vogt KC, Aquino N, Younes A, Scheinberg DA: Potentiating antibody-dependent killing of cancers with CAR T cells secreting CD47-SIRPalpha checkpoint blocker. Blood 2023;141:2003-2015.
https://doi.org/10.1182/blood.2022016101 |