A Comprehensive Pan-Cancer Analysis of the Mitochondrial Uncoupling Protein UCP2, with a Focus on Sex and Gender-Related Aspects
Keywords
Abstract
Background/Aims:
Mitochondrial uncoupling protein 2 (UCP2) plays a crucial role in regulating oxidative stress and cellular metabolism, positioning it as an important subject in oncological research. The involvement of UCP2 in cancer is complex and context-dependent, suggesting it as a potential therapeutic target. In this study, we aimed to perform a comprehensive pan-cancer analysis of UCP2, with a particular focus on gender-related malignancies such as breast (BRCA), prostate (PRAD), ovarian (OV), and testicular tumors (TGCT).Methods:
We analyzed UCP2 expression in The Cancer Genome Atlas (TCGA), examining correlations with prognosis, tumor mutational burden (TMB), microsatellite instability (MSI), immune cell infiltration, immune checkpoint genes, genes involved in steroidogenesis, sex hormone receptor genes, and drug sensitivity.Results:
Significant variability in UCP2 expression was observed across cancer types. UCP2 levels were elevated in BRCA and OV but reduced in PRAD and TGCT. High UCP2 expression was associated with a better prognosis in OV and poorer overall survival in PRAD. Furthermore, UCP2 correlated with TMB and MSI in OV, TGCT, and BRCA. UCP2 expression was also linked to immune cell infiltration, immune checkpoint genes, steroidogenic genes, and sex hormone receptor genes, with variable effects depending on cancer type and gender. Additionally, UCP2 also demonstrated sensitivity to specific anticancer drugs.Conclusion:
Our findings highlight the interplay between UCP2, immune and hormonal pathways, and drug response and reveal potential opportunities for new therapeutic combinations, especially in gender-related cancers.Introduction
Cancer development involves unique metabolic changes, now considered hallmarks of carcinogenesis [1]. Otto Warburg was the first to observe that cancer cells, even in the presence of oxygen rely mainly on glycolysis, rather than oxidative phosphorylation (OXPHOS), for energy production. This metabolic rewiring, known as the “Warburg effect,” was initially attributed to reduced mitochondrial activity in cancer cells. Additional studies have demonstrated that many cancer cells have intact mitochondria, crucial for their metabolism and various cellular processes, including proliferation, survival in a challenging microenvironment, and metastasis [2, 3]. Mitochondria are essential for providing eukaryotic cells with energy in the form of ATP [4]. The metabolic breakdown of nutrients generates electrons, which are transferred through the electron transport chain (ETC) located in the inner mitochondrial membrane (IMM), generating a proton gradient across the IMM and establishing the mitochondrial potential essential for ATP synthase to produce ATP from ADP and inorganic phosphate [5]. In addition to ATP production, the ETC also generates reactive oxygen species (ROS), which play a role in homeostatic signaling but can also cause oxidative stress in disease conditions [6]. The ETC is vulnerable to ROS production under conditions of high electron flow and elevated mitochondrial membrane potential. Thus, uncoupling oxidative phosphorylation (OXPHOS) can reduce respiration rate and decrease mitochondrial ROS production. Physiologically, mitochondrial OXPHOS is not fully coupled to ATP production. Protons can leak back into the mitochondrial matrix from the IMS, bypassing ATP synthase and generating heat instead of ATP. These events can be regulated by uncoupling proteins (UCPs), a family of proton transporters located in the IMM [7],[8]. Among the UCPs, UCP2 is the most ubiquitously expressed, found in multiple cell types. The role of UCP2 in ATP production has been extensively investigated in numerous studies, particularly in pancreatic β-cells, identifying UCP2 as a negative regulator of ATP levels. By promoting proton leakage across the IMM, UCP2 lowers the mitochondrial membrane potential and reduces ATP synthesis [9, 10, 11, 12].
UCP2 has been identified as a crucial regulator of ROS in various cell types. It functions by mildly uncoupling OXPHOS, which reduces the mitochondrial membrane potential and ROS production. This activity provides an antioxidant effect, which helps to protect cells from oxidative damage. However, the role of UCP2 in uncoupling OXPHOS and its impact on ROS regulation remains under debate [13, 14, 15, 16, 17, 18, 19]. In addition, UCP2 emerged as a crucial regulator of cellular metabolism [20]. Therefore, several research studies acknowledged its involvement in various physiological and pathological conditions [21, 22]. In physiological conditions, UCP2 exerts tissue-specific roles essential to protecting cells from oxidative stress, maintaining metabolic homeostasis, and regulating immune responses. Its dysregulation is linked to a range of pathological conditions, including metabolic disorders and cancer. While UCP2 has been extensively studied in cancer in recent years, its exact role in carcinogenesis remains ambiguous. UCP2 is often highly expressed in cancer. Several cancer types have established a correlation between elevated UCP2 expression and tumor progression, highlighting its potential as a therapeutic target to treat UCP2 overexpressing cancers [23, 24]. Conversely, other studies have shown that UCP2 overexpression reduces tumorigenicity, alters mitochondrial signaling, and enhances chemotherapeutic-induced apoptosis by acting as a metabolic tumor suppressor [25, 26]. Interestingly, as a metabolic sensor, UCP2 regulates mitochondrial substrate oxidation by modulating the exchange of critical metabolites, influencing cellular energy balance and metabolic pathways in response to varying nutrient availability [27]. Therefore, UCP2 is implicated in the metabolic adaptations of cancer cells beyond its traditional uncoupling [28, 29, 30].
The tumor microenvironment (TME) immune composition and the immune profile of patients are increasingly recognized as critical determinants of cancer progression, therapeutic response, and prognosis [31, 32, 33, 34]. UCP2 intrinsically modulates tumor cell proliferation and chemoresistance, influencing their ability to evade immunosurveillance [35]. Furthermore, UCP2 is expressed in immune cells, including macrophages [36]. As major immune infiltrates in solid tumors, macrophages adapt to the hypoxic tumor environment by reprogramming their metabolism in an activation state that promotes tumor growth [37]. In the TME, macrophages adopt distinct activation states and metabolic profiles. Pro-inflammatory (M1) macrophages, characterized by glycolytic metabolism, exhibit anti-tumorigenic properties, while anti-inflammatory (M2) macrophages, which rely on OXPHOS, display protumor activities [38]. UCP2 is involved in the polarization and metabolism of macrophages. During M1 polarization, UCP2 is downregulated, while its overexpression decreases macrophage ROS production, promotes metabolic rewiring towards OXPHOS, and re-polarization from M1 to M2 phenotype [39]. Therefore, UCP2 is an exciting target for understanding the molecular mechanisms behind metabolic reprogramming and macrophage activation in cancer. Further studies are necessary to clarify the role of UCP2 in macrophages in cancer development and progression. More recently, UCP2 has been suggested to play a role in steroidogenesis in vitro and in vivo, specifically affecting sex hormone production. More recently, UCP2 has been suggested to play a role in steroidogenesis in vitro and in vivo, specifically affecting sex hormone production [40]. Additionally, there is evidence to indicate that sex hormones could influence the expression of UCP2 [41, 42], which has led to speculations that UCP2 might be involved in regulating gender-specific variations in disease responses, including cancer.
In this study, we conducted a comprehensive pan-cancer analysis of UCP2 using data from The Cancer Genome Atlas (TCGA). Our investigation focused on UCP2 expression patterns, prognostic significance, genetic alterations, tumor mutational burden (TMB), microsatellite instability (MSI), immune infiltration, and the involvement of genes related to immunotherapy, steroidogenesis, and sex hormone receptors in the TCGA dataset. Special emphasis was placed on gender-related cancers, specifically breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers (Fig. 1).
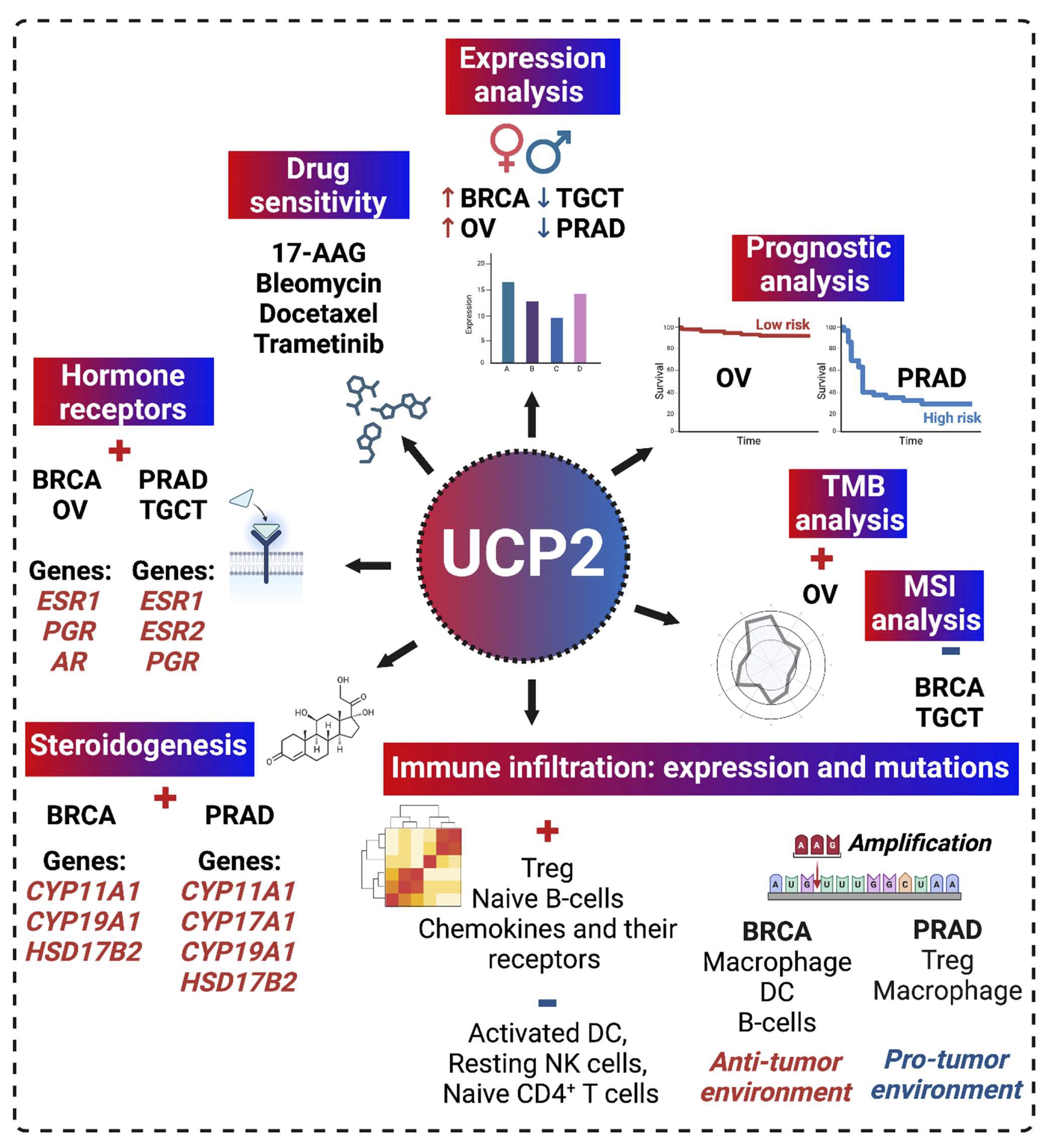
Fig. 1: Graphical abstract. The graphical abstract illustrates the methodology and key findings, including UCP2 expression, prognosis, correlations with TMB, MSI, immune infiltration, steroidogenic genes, sex hormone receptor genes, and drug sensitivity in BRCA, PRAD, OV, and TGCT cancer types.
Materials and Methods
Data processing
The TCGAbiolinks R package (v2.32.0) along with BiocManager (v1.30.23) were employed to download TPM matrix, clinical information, TMB, and methylation matrices for the 33 types of cancers available in TCGA (https://portal.gdc.cancer.gov/). This was achieved using the functions GDCquery, GDCdownload, and GDCprepare. The expression matrix was generated using the formula log2(TPM+1) and prepared using the TCGAplot R package (v7.0.1)[43]. No data quality checks were performed. The ggpubr R package (v0.6.0) was used to compare and plot UCP2 expression across different tissues.
AUC analysis
TCGA tumors with enough normal samples were selected for Area Under the Curve (AUC) analysis based on UCP2 expression. The pROC R package (v1.18.5) was used to assess the diagnostic potential of UCP2 in distinguishing tumor samples from normal samples. The ROC curve was plotted, and the AUC and the 95% confidence interval (CI) were presented.
Prognosis potential analysis of UCP2
Overall Survival (OS) was evaluated to assess the predictive potential of UCP2. The survminer (v0.4.9) and survival (v3.6-4) R packages were used to perform the log-rank test and to generate Kaplan-Meier (KM) plots for data grouped by UCP2 expression. The survival and forestplot (v3.1.3) R packages were employed to conduct Cox proportional hazards analysis to determine the relationship between UCP2 expression and survival across 33 types of cancers in TCGA.
Correlation of UCP2 expression with mutational tumor heterogeneity
The correlation between UCP2 expression and Tumor Mutational Burden (TMB) or Microsatellite Instability (MSI) was assessed using Pearson correlation analysis. The radar plot illustrating these correlations was generated using the fmsb R package (v0.7.6).
Relationship between UCP2 expression and immunity
A list of immune-related genes was downloaded from TISIDB (http://cis.hku.hk/TISIDB/index.php) [44]. The correlation between UCP2 expression and immune-related genes and immune infiltration was examined using Pearson correlation analysis. The results were visualized using the ggplot2 R package (v3.5.1). The estimate R package (v1.0.13) was used to compute each specimen's immune, stromal, and estimate scores for each specimen. Pearson correlation analysis assessed the relationship between UCP2 expression and these scores. The correlations were plotted using the ggplot2 (v3.5.1) and ComplexHeatmap (v2.20.0) R packages.
Biological significance of UCP2 in tumors
Cancer specimens were divided into UCP2 high and low-expression subgroups. Differentially regulated genes between these subgroups were identified using the limma R package (v3.60.4). All genes were ranked by fold change (high-risk vs low-risk group) and used as input for GSEA-GO and GSEA-KEGG analyses. These analyses were performed using the org.Hs.eg.db (v3.19.1) and clusterProfiler (v4.12.1) R packages, with data from the KEGG database (https://www.kegg.jp/kegg/). The top five terms were visualized using the enrichplot R package (v1.24.2).
Drug sensitivity analysis
Gene Set Cancer Analysis (GSCA, http://bioinfo.life.hust.edu.cn/GSCA/#/, accessed on 29 July 2024) is an integrated genomic, pharmacogenomic, and immunogenomic gene set cancer analysis platform. This platform was utilized for drug sensitivity analysis related to UCP2 expression.
RNA expression in single-cell clusters
UMAP plots were generated to visualize the human expression of UCP2 in single cells across BC, OV, PRAD, and TGCT cancers.
Results
Expression and prognostic analysis of UCP2 in human cancers
The expression of UCP2 was evaluated across various tumor types using data from the TCGA database (Table S1). When comparing the expression of UCP2 between tumor and normal tissues, we found a significant increase in 9 types of tumors, including bladder cancer (BLCA), breast cancer (BRCA), cholangiocarcinoma (CHOL), esophageal cancer (ESCA), glioblastoma multiforme (GBM), lung adenocarcinoma (LUAD), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) (Fig. 2A, Table S2). In contrast, reduced UCP2 expression was observed in kidney chromophobe (KICH), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), and prostate adenocarcinoma (PRAD) (Fig. 2A, Table S2). We were unable to detect significant differences in UCP2 expression in BRCA-Basal, BRCA-Her2, BRCA-LumA, BRCA-LumB, diffuse large B-cell lymphoma (DLBC), acute myeloid leukemia (LAML), low-grade glioma (LGC), mesothelioma (MESO), ovarian cancer (OV), sarcoma (SARC), testicular germ cell tumors (TGCT), uterine carcinosarcoma (UCS), and uveal melanoma (UVM) due to the lack of normal tissue data for comparison (Fig. 2A, Table S2). We then investigated the levels of UCP2 expression in normal tissues and cancer cell lines using data from the Human Protein Atlas (HPA). In normal tissues, higher expression of UCP2 was found in lymphoid organs, followed by the female reproductive system. In contrast, low expression was observed in the male reproductive system and most brain regions (Fig. S1A). Additionally, higher UCP2 expression was identified in macrophages of adipose and liver tissues (Fig. S1B). Among cancer cell lines, the highest UCP2 expression was found in lymphoma cells, followed by leukemia, adrenocortical, and breast cancer cell lines (Fig. S1C).
Next, we examined the predictive significance of UCP2 expression on cancer patient survival using the Pan-cancer Cox proportional hazards model (HR) (Fig. 2B, C; Table S3). The overall survival (OS) results revealed that UCP2 expression is a risk factor in kidney renal papillary cell carcinoma (KIRP) (HR = 1.336, p = 0.018), acute myeloid leukemia (LAML) (HR = 1.921, p = 0.0002), and brain lower grade glioma (LGG) (HR = 1.392, p = 0.0002). Conversely, it was associated with improved survival outcomes in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (HR = 0.719, p = 0.004), lung adenocarcinoma (LUAD) (HR = 0.766, p = 0.008), ovarian serous cystadenocarcinoma (OV) (HR = 0.849, p = 0.019), and sarcoma (SARC) (HR = 0.782, p = 0.011) (Fig. 2B, Table S3). The age-adjusted hazard ratio analysis revealed that UCP2 expression is a risk factor in glioblastoma multiforme (GBM) (HR = 1.278, p = 0.027), kidney renal papillary cell carcinoma (KIRP) (HR = 1.384, p = 0.014), acute myeloid leukemia (LAML) (HR = 1.744, p = 0.001), and brain lower grade glioma (LGG) (HR = 1.599, p = 0.0002) (Fig. 2C, Table S4). Additionally, it suggested that UCP2 is a protective factor in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (HR = 0.729, p = 0.006), ovarian serous cystadenocarcinoma (OV) (HR = 0.871, p = 0.051), lung adenocarcinoma (LUAD) (HR = 0.727, p = 0.002), and sarcoma (SARC) (HR = 0.742, p = 0.003) (Fig. 2C, Table S4). These results indicate that UCP2 levels vary among different cancer types, showing elevated expression in certain tumors, including breast cancer, and decreased levels in others, such as prostate cancer. It represents a risk factor in some tumors with downregulated UCP2 levels while correlating with beneficial outcomes in others with increased UCP2 levels, such as ovarian cancer.
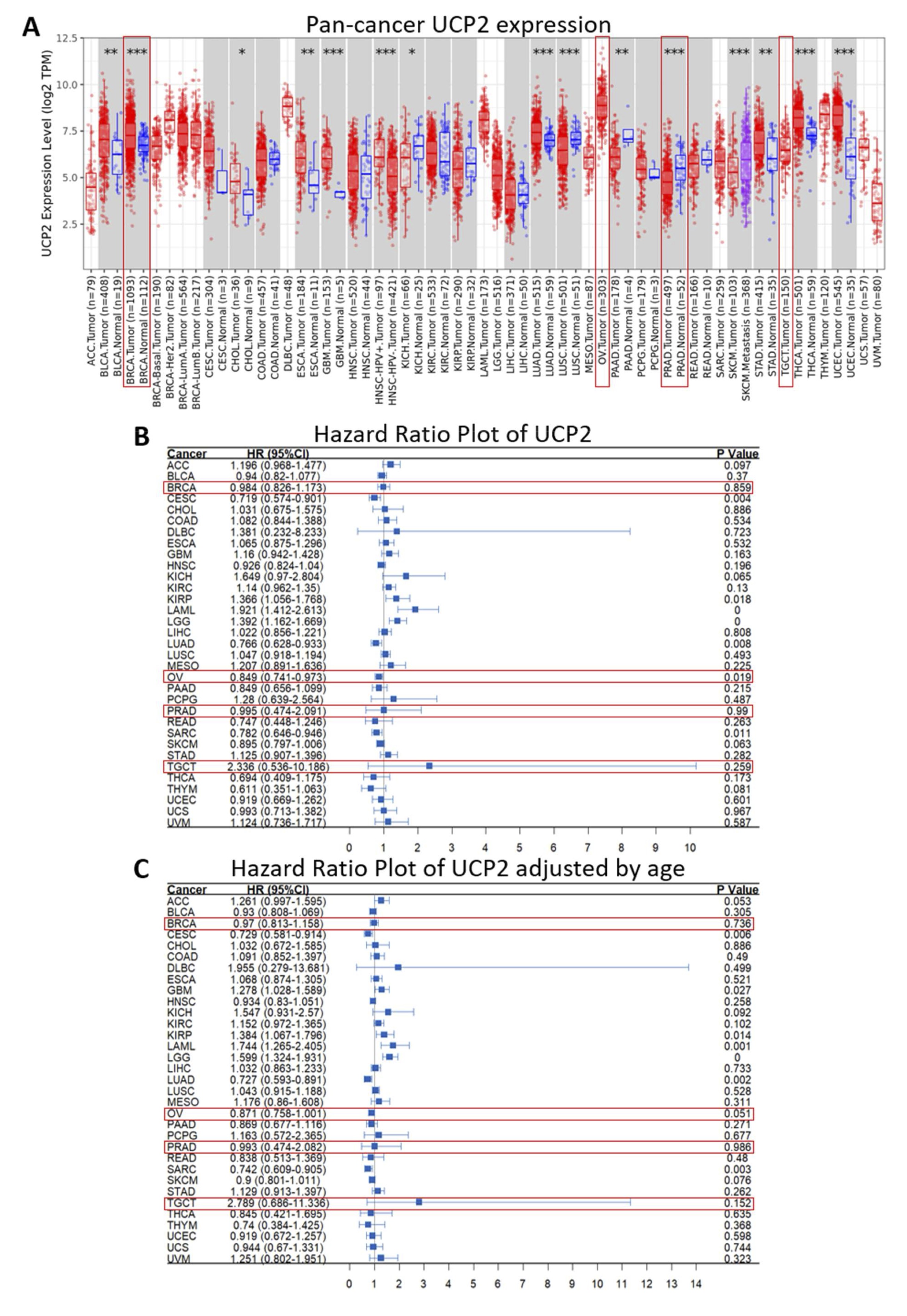
Fig. 2: Expression patterns of UCP2 across various cancer types. (A) TIMER2.0 analysis illustrating UCP2 expression levels in TCGA cancers compared to normal tissues, with significance denoted as ***p < 0.001; **p < 0.01; *p < 0.05. Pan-cancer risk analysis. Pan-cancer Cox proportional hazards model analysis of UCP2 across 33 types of tumors in TCGA. The Wilcoxon test was used to compare UCP2 expression between normal and tumor groups, with significance indicated as ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, and ns denoting no significant difference. The function provides the ability to perform pan-cancer Cox regression analysis without (B) or with age adjustment (C), visualized by a forest plot.
Genomic mutations, TMB, and MSI analysis of UCP2 in pan-cancer
We used the cBioPortal Database to examine UCP2 genetic variations in the TCGA pan-cancer cohort. UCP2 genetic alterations, including amplifications, mutations, and deep deletions, were observed in many cancers. UCP2 amplifications were most common in ovarian (OV) and breast (BRCA) cancers. Meanwhile, mutations were most prevalent in kidney chromophobe (KICH), and deep deletions were characteristic of testicular germ cell tumors (TGCT) (Fig. 3A).
Tumor mutational burden (TMB) and microsatellite instability (MSI) are molecular markers associated with tumor immunogenicity and can be used as potential biomarkers to predict responses to immunotherapy [45, 46]. We conducted a Pearson correlation analysis to explore the connection between UCP2 expression and these markers. Our research uncovered a significant positive correlation between UCP2 expression and TMB in ovarian cancer (OV), and a negative correlation was found in kidney renal papillary cell carcinoma (KIRP) (Fig. 3B). UCP2 was negatively linked with MSI in testicular germ cell tumors (TGCT), breast invasive carcinoma (BRCA), KIRP, lung squamous cell carcinoma (LUSC), and stomach adenocarcinoma (STAD), while it showed a positive correlation exclusively in lung adenocarcinoma (LUAD) (Fig. 3C). In conclusion, considering gender dependent cancers, these results indicate that UCP2 amplifications are frequent in OV while deletions occur in TGCT, and UCP2 may affect antitumor immunity through its association with TMB and MSI in these cancers.
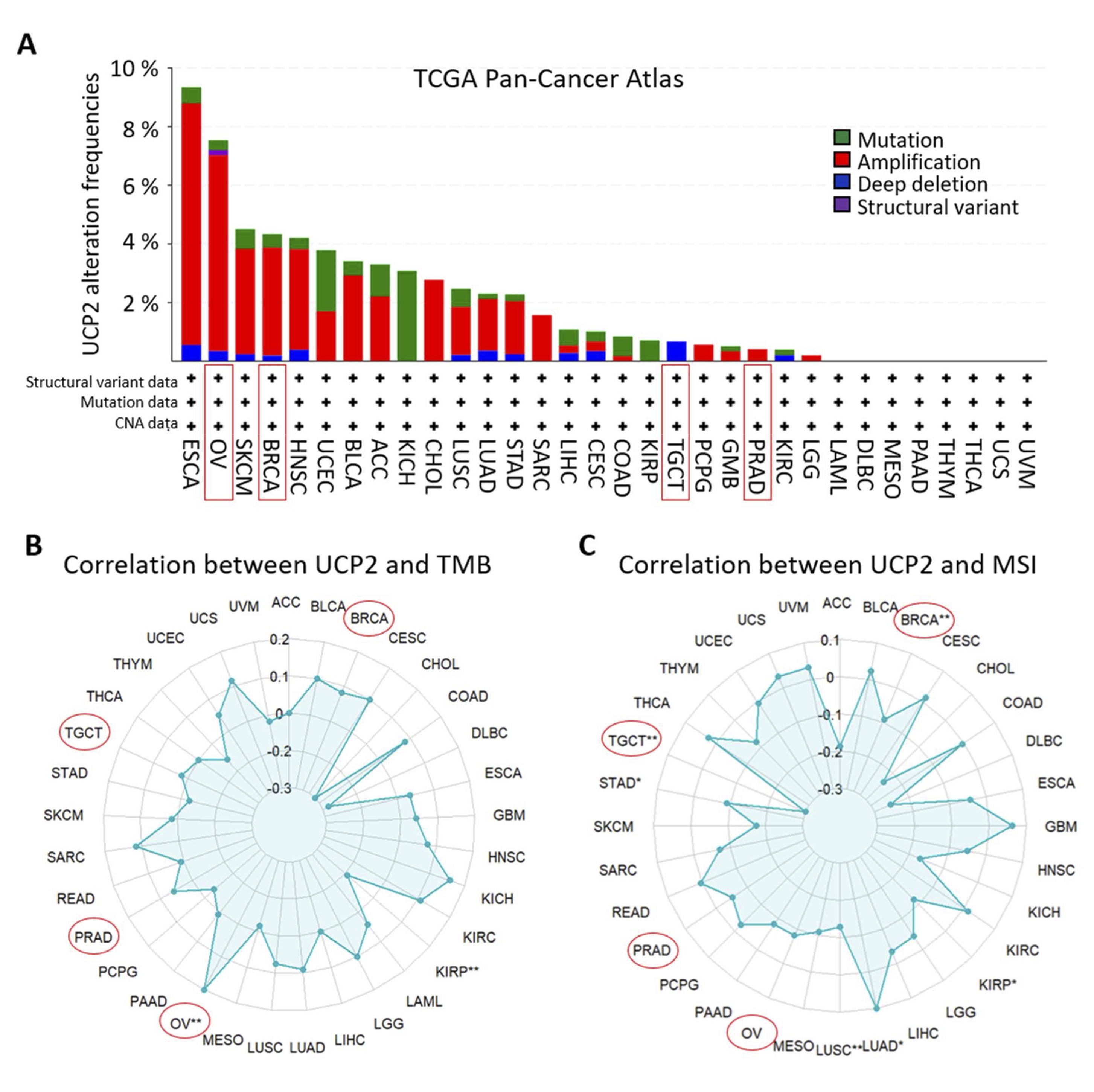
Fig. 3: Mutation analysis of UCP2, correlation with TMB and MSI in pan-cancer. (A) Analysis using cBioPortal revealed the mutation types, frequencies, and sites of UCP2 across various cancer types. (B) Pan-cancer correlation analysis of UCP2 with Tumor Mutational Burden (TMB). (C) Pearson correlation analysis was used to calculate the correlation between UCP2 expression and MSI, with significance indicated as **p < 0.01 and *p < 0.05. They were generated using the TCGAplot R package (v7.0.1) (https://github.com/tjhwangxiong/TCGAplot).
Correlation analysis of UCP2 expression with immune cell infiltration and checkpoint genes in pan-cancer
Considering the crucial role of the tumor immune microenvironment, its impact on clinical outcomes [47], and the established involvement of UCP2 in the immune system [48], we investigated the correlation between UCP2 expression and immune cell infiltration in human cancers. Significant positive correlations between UCP2 and stromal, immune, and estimate scores were found in almost all cancers. In contrast, only a few types of tumors, including ovarian cancer, showed no correlations with immune characteristics (Fig. 4A). Furthermore, in most tumor samples from the 33 tumor types, UCP2 expression was positively correlated with regulatory T cells (Tregs) and naive B cells, while showing negative associations with activated dendritic cells (DC), resting NK cells, and naive CD4+ T cells (Fig. 4B). Targeting tumor immune infiltrates alone or in combination with immunotherapy can improve patient outcomes [38]. Therefore, we evaluated the correlation between UCP2 gene expression and immune checkpoint-associated genes in cancers from the TCGA. UCP2 positively correlated with most immune checkpoint genes in nearly all analyzed tumors. Particularly strong correlations were evident in testicular and prostate cancers. However, moderate associations were found in breast cancer. Interestingly, no significant correlations were observed in ovarian cancer, suggesting that UCP2 may not play a key role in immune regulation within the tumor microenvironment of this cancer type (Fig. 4C).
Additionally, we investigated the association of UCP2 with the immune inhibitory (Fig. 4D) and immune stimulatory genes (Fig. 4E), chemokine-related genes, and chemokine receptor genes (Fig. S2A, B). Interestingly, UCP2 showed strong positive correlations with most immune inhibitory and stimulatory genes in cancers with higher incidence in males, such as kidney renal papillary cell carcinoma (KIRP), kidney renal clear cell carcinoma (KIRC), head and neck squamous cell carcinoma (HNSC), and liver hepatocellular carcinoma (LIHC) (Fig. 4D, E; Table S5). In contrast, the most pronounced negative correlations were observed in cancers such as cervical squamous cell carcinoma (CESC), uterine corpus endometrial carcinoma (UCEC), lung adenocarcinoma (LUAD), and thymoma (THYM) (Fig. 4D, E; Table S5). Accordingly, strong positive correlations between UCP2 and these genes were found in prostate and testicular cancers, while in breast and ovarian cancer, the correlations were weak or absent. These results suggest that UCP2 may have varying effects on immune characteristics depending on the cancer type and potentially showing a sex-biased incidence.
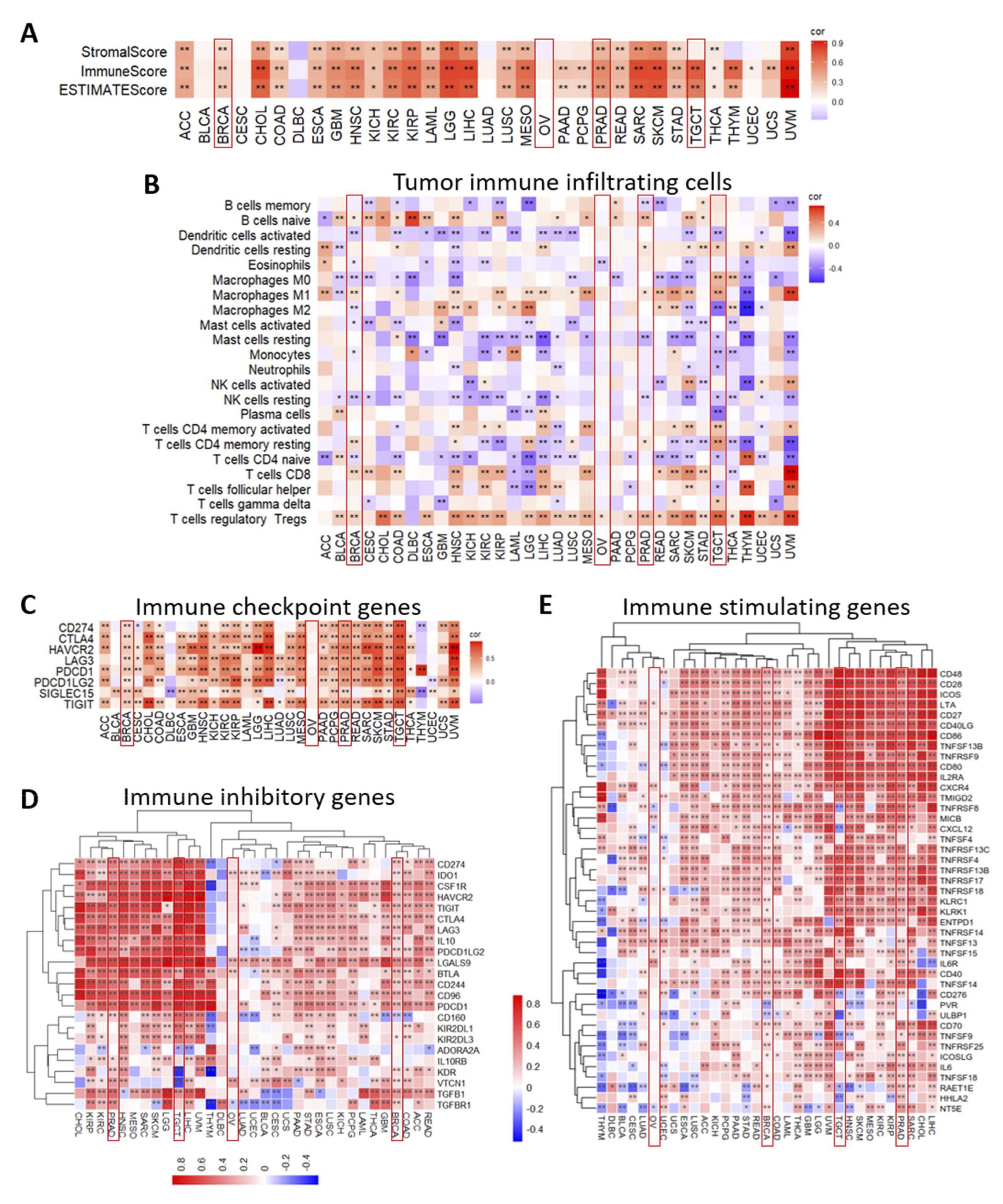
Fig. 4: Correlation between UCP2 expression and immune properties in pan-cancer. (A) Heatmap showing pan-cancer correlation analysis of UCP2 and immune scores, generated using Pearson correlation analysis. (B) Correlation analysis between UCP2 expression and tumor-infiltrating immune cells (TIICs) in 33 cancer types. Pearson correlation analysis was used to calculate the correlation between UCP2 expression and immune cell ratio. (C) Immune checkpoint-associated genes. (D) Immune inhibitory genes. (E) Immune stimulating genes. Similar clusters were identified using the complete linkage method, with significance levels indicated as **p < 0.01 and *p < 0.05. The heatmaps were drawn using the TCGAplot R package (v7.0.1) (https://github.com/tjhwangxiong/TCGAplot). Pearson correlation analysis was used to calculate the correlation between UCP2 expression and these immune-related genes.
Correlation analysis of UCP2 expression with steroidogenic and sex hormone receptor genes in gender-related cancers
Sex steroids play crucial roles in various biological processes. However, they can also promote cancer-related characteristics by affecting cell proliferation, migration, and invasiveness. Alterations or malfunctions in the steroidogenic pathway may contribute to the development of cancers, such as breast, ovarian, prostate, and testicular cancer [49, 50, 51, 52, 53]. UCP2 may impact steroid production by affecting mitochondrial function and cellular metabolism [54, 55]. We examined the connection between UCP2 and several key genes involved in steroidogenesis (Fig. 5A) and identified significant correlations with essential enzymes in sex steroid biosynthesis [56]. In breast cancer (BRCA) and prostate cancer (PRAD), UCP2 was found to be associated with CYP11A1, CYP19A1, and HSD17B2. A positive correlation between UCP2 and CYP17A1 was also observed in PRAD and testicular germ cell tumors (TGCT). Furthermore, UCP2 showed a positive correlation with HSD3B7 in ovarian cancer (OV) and BRCA (Fig. 5A). Sex steroids exert their biological functions by interacting with specific sex hormone receptors, including estrogen receptors (ESR), progesterone receptors (PGR), and androgen receptors (AR) [57]. Thus, we investigated the correlation between UCP2 and these genes (Fig. 5B). In BRCA, UCP2 exhibited slight but statistically significant positive correlations with ESR1 (R=0.082, p=0.006), PGR (R=0.08, p=0.0076), and with AR (R=0.27, p<2.2e−16) (Fig. 5C). In OV, UCP2 was linked with ESR1 (R=0.21, p=1.3e−05). (Fig. 5C). In both PRAD and TGCT, UCP2 was associated with ESR1 (PRAD: R=0.32, p<0.05; TGCT: R=0.36, p=4.1e−06), ESR2 (PRAD: R=0.15, p<0.05; TGCT: R=0.24, p=0.0025), and PGR (PRAD: R=0.26, p=2.2e−09; TGCT: R=0.25, p=0.0017) but not with AR (Fig. 5C). These data indicate that UCP2 expression widely correlates with key steroidogenic genes in cancers and may influence sex hormone pathways by interacting with sex receptor genes.
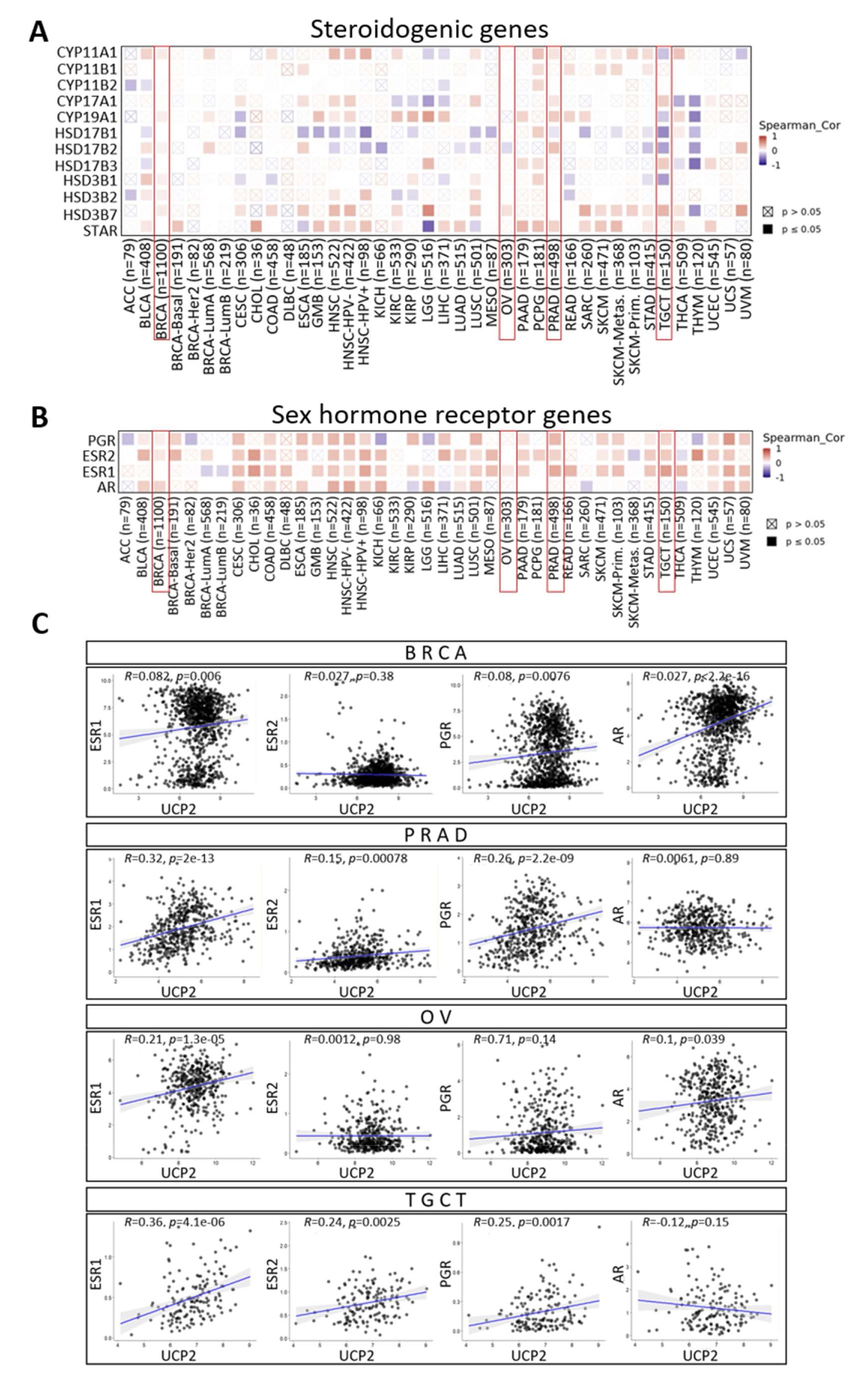
Fig. 5: Correlation analysis of UCP2 expression with steroidogenic and sex hormone receptor genes. Heatmaps from TIMER2.0 analysis showing pan-cancer correlation analysis of UCP2 and genes related to steroidogenesis (A) and sex hormone receptors (B). The color coding represents the strength and direction of the correlations, with red indicating a positive correlation, blue indicating a negative correlation, and white indicating no correlation. Grey crosses denote non-significant correlations (p > 0.05). (C) Correlation Plots of UCP2 Expression with hormone receptor genes (ESR1, ESR2, PGR, AR) Across Breast, Prostate, Ovarian, and Testicular Cancers by using the TCGAplot R package (v7.0.1) (https://github.com/tjhwangxiong/TCGAplot)
The multifaceted role of UCP2 in tumor progression and immune modulation in breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers
Considering the results showing alterations of UCP2 expression in various tumor types, its associations with immune characteristics, and its potential involvement in hormone-driven cancers, we further explored its role in breast, prostate, ovarian, and testicular malignancies. Genetic and epigenetic alterations are crucial in tumorigenesis, affecting gene expressions and interfering with key cellular pathways that regulate cellular proliferation and functions [58]. Our analysis revealed distinct differences in the copy number alterations (CNA) of the UCP2 gene between female and male cancers. A high amplification rate of UCP2 was found in BRCA and OV, while deletions were mainly observed in TGCT. PRAD showed relatively low levels of both amplifications and deletions (Fig. 6A, B). Furthermore, we investigated the promoter methylation of UCP2 and revealed significant inverse correlations with UCP2 expression in BRCA, PRAD, and TGCT. High promoter methylations correlated with reduced UCP2 expression in these cancers (Fig. 6C). The analysis of copy number variations (CNVs) revealed that amplifications and deletions of UCP2 affected immune cell infiltration (Fig. 6D). In BRCA, amplifications were associated with an increase in B cells, dendritic (DC) cells, and macrophages, as well as with a decrease in CD4 T and natural killer (NK) cells. On the other hand, deletions were linked to an increment in CD8 naive T cells. In OV, amplifications enhanced induced regulatory T cells (iTreg), while deletions, like in breast cancer, increased CD8 naive cells. In PRAD, amplifications increased macrophages, Th1 cells, DC cells, and iTreg cells. Conversely, deletions increased cytotoxic and NK cells but reduced CD4 naive and Th17 cells. In TGCT, deletions, like in PRAD, increased cytotoxic cells (Fig. 6D; Supplementary file 1). UCP2 is highly expressed in macrophages, predominant immune cells in breast, prostate, ovarian, and testicular tissues (Fig. S3). Therefore, we aimed to investigate whether variations in the copy number of UCP2 could impact macrophage infiltration in these cancer types (Fig. 6E). UCP2 amplifications were correlated with increased macrophage infiltration in BRCA and PRAD but not in OV and TGCT. These data indicate that methylation and copy number alterations (CNAs) impact the expression of UCP2 and influence the tumor microenvironment (TME). Amplifications in female cancers (BRCA and OV) promote immune cell infiltration and immunosuppressive profiles, while deletions in male cancers (PRAD and TGCT) favor cytotoxic immune responses.
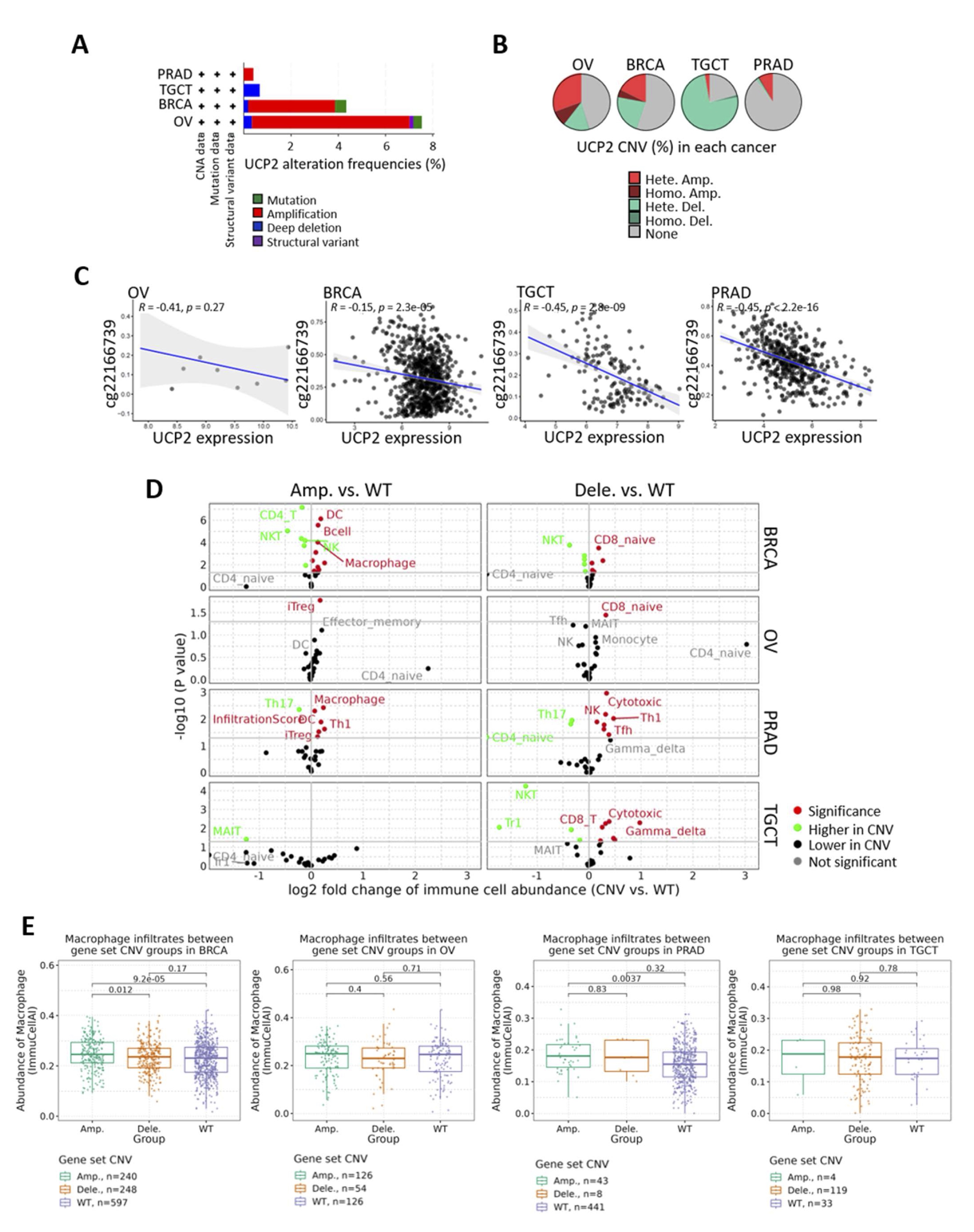
Fig. 6: Mutation and methylation analyses of UCP2 across BRCA, OV, PRAD, and TGCT tumors. (A) Mutation types, frequencies, and sites of UCP2 across cancer types from the cBioPortal. (B) The Gene Set Cancer Analysis (GSCA, http://bioinfo.life.hust.edu.cn/GSCA/#/) was used to define copy number variations (CNVs). (C) Scatter methylation plot (Scatter Pie Plot v0.2.3) showing the correlation between UCP2 expression and promoter methylation in BRCA, PRAD, OV, and TGCT by using the TCGAplot R package (v7.0.1) (https://github.com/tjhwangxiong/TCGAplot). (D) A volcano plot was generated using GSCA, showing the log2 fold change in immune cell abundance between CNV and WT samples. (E) Box plot from GSCA investigating the CNV of UCP2 and macrophage infiltration in BRCA, PRAD, OV, and TGCT.
Impact of UCP2 expression and methylation on overall survival in breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers
Given the observed impact of UCP2 alterations on tumor immune microenvironments, we evaluated how UCP2 expression (Fig. 7A) and methylation signature (Fig. 7B) affect overall survival in breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers. Several lines of research investigated the UCP2 impact on patient survival in breast cancer. Some studies indicated that higher UCP2 expression was associated with a worse prognosis [59], while others have revealed increased survival [60]. Our data aligns with the studies [61], showing no significant impact of UCP2 expression on BRCA survival (Fig. 7A). We observed that increased UCP2 expression correlated with prolonged survival in OV (p=0.043), while it was associated with decreased survival in PRAD (p=0.032) (Fig. 7A). DNA methylation, a critical epigenetic mechanism influencing cancer development and progression, also contributes to patient survival [62]. High DNA methylation at cg22166739UCP2 was associated with decreased UCP2 expression and was linked to increased overall survival in BRCA patients (p=0.0087). The methylation level of UCP2 did not affect survival outcomes in OV, PRAD, and TGCT patients (Fig. 7B). To further understand the complex role of UCP2 and its impact on patient prognosis in these cancers, we conducted an analysis of patient survival using the Kaplan-Meier survival and COX proportional hazards model based on the cancer stage. Considering this variable, high UCP2 expression was associated with reduced survival in advanced BRCA and TGCT cancer stages (Fig. S4A-D). These results indicate that UCP2 expression and methylation impact prognostic outcomes, which vary depending on the type of cancer and possible gender-related discrepancies.
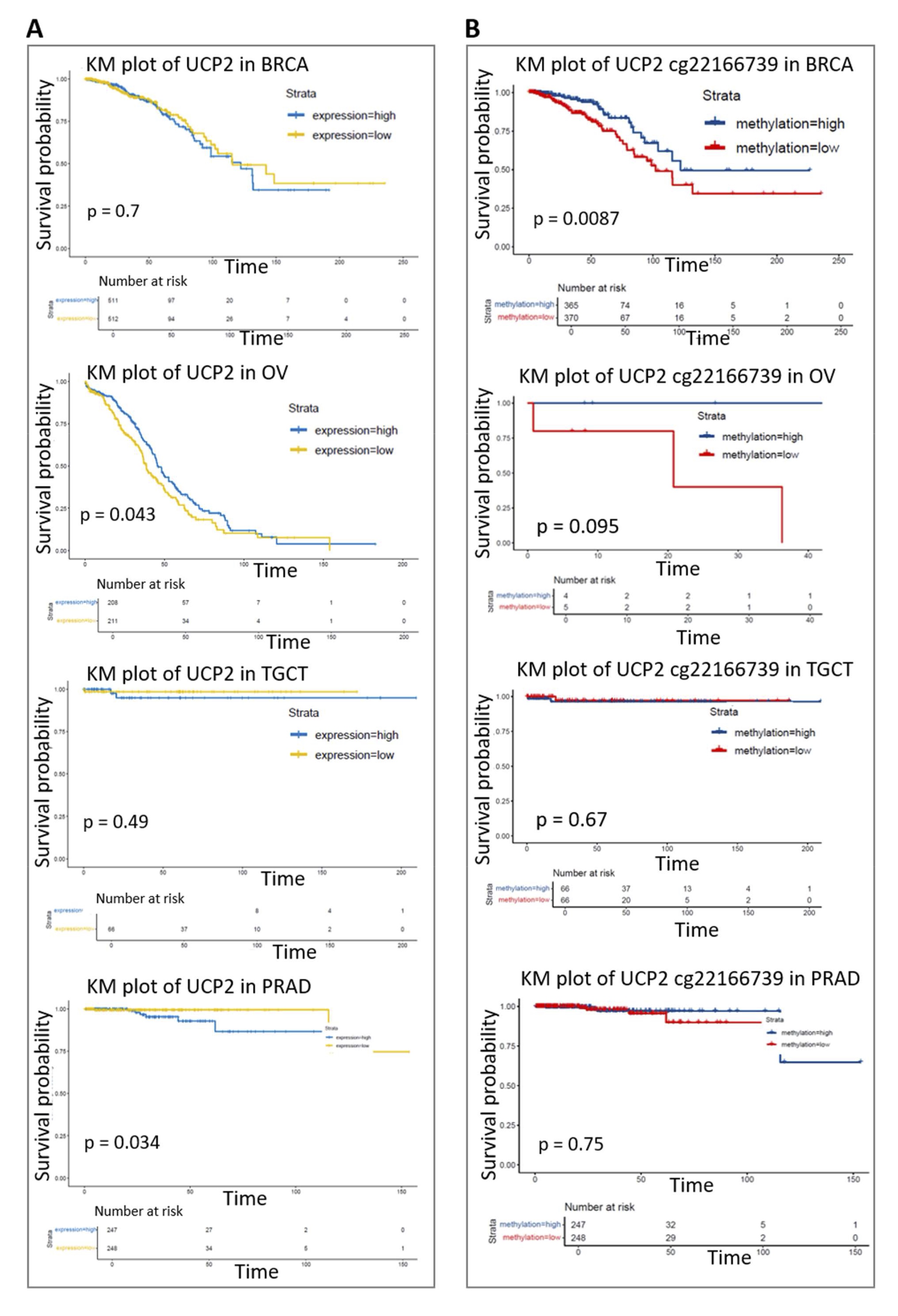
Fig. 7: Kaplan-Meier (KM) analysis. (A) KM analysis examining the correlation between UCP2 expression and overall survival (OS) in patients with BRCA, PRAD, OV, and TGCT. (B) KM analysis investigating the correlation between UCP2 methylation and OS in BRCA, PRAD, OV, and TGCT patients. The KM plots were generated using the TCGAplot R package (v7.0.1) (https://github.com/tjhwangxiong/TCGAplot).
Correlation analysis of UCP2 and clinical phenotypes in breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers
Patients over 60 showed significantly lower UCP2 expression in OV. Still, no such reduction was observed in BRCA, PRAD, or TGCT (Fig. 8A). The elevated UCP2 expression in younger OV patients suggests a potential role for UCP2 in the early onset or more aggressive forms of the disease. We also evaluated the expression difference of UCP2 in various tumor stages (Fig. 8B). The analysis did not reveal statistically significant differences in UCP2 expression across tumor stages of these cancers. However, the available data was insufficient for a conclusive study. In OV, a notable trend of increased UCP2 expression was observed in the early stages, suggesting that UCP2 may play a role in the initial phases of tumor development. Conversely, a tendency toward decreased UCP2 expression in advanced stages of TGCT may indicate a role in tumor suppression as the disease progresses (Fig. 8B). The expression of UCP2 and its relationship with metastasis varies across different cancer types (Fig. 8C). In BRCA and OV, UCP2 expression is higher in primary tumor tissues compared to normal tissues. However, its expression decreases during metastasis, although it remains higher than in normal tissues. This indicates that UCP2 may play a dynamic role in early tumor progression but may become less involved as metastasis advances. The expression of UCP2 in PRAD tumors is lower in primary tumors compared to normal tissues and even more reduced in metastatic tumors, suggesting that UCP2 may play a suppressive role in the progression of prostate cancer (Fig. 8C). For TGCT, the available data was insufficient to draw definitive conclusions about UCP2 expression patterns in metastasis (Fig. 8C). These results indicate that UCP2 levels are higher in primary BRCA and OV tumors but lower in primary tumors of PRAD. UCP2 levels decrease during metastasis in both female (BRCA, OV) and male (PRAD) cancers, although this decline is more consistent in PRAD.
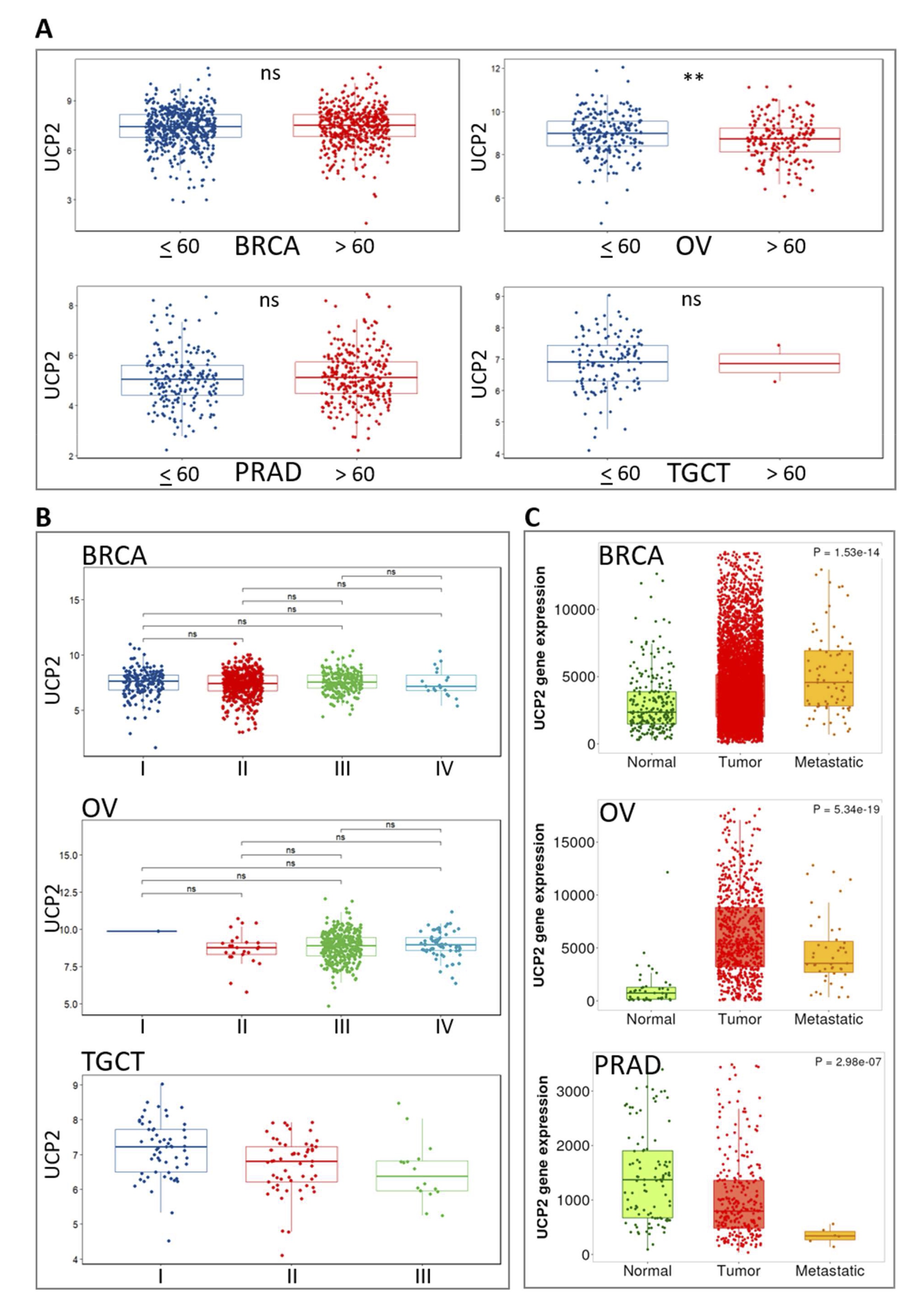
Fig. 8: Pan-cancer correlation analysis of UCP2 and clinical phenotypes. (A) Expression of UCP2 in patients aged ≤ 60 and > 60 years in BRCA, PRAD, OV, and TGCT. The Wilcoxon test was used for comparisons. (B) Correlation of UCP2 expression with BRCA, OV, and TGCT tumor stage. Data is shown as log2(TPM + 1), and the Wilcoxon test was used to compare different groups. ns indicates no significant difference. (C) Correlation of UCP2 expression with primary and metastatic tumors in BRCA, PRAD, and OV.
The potential functions of UCP2 in breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers
We examined the correlation between UCP2 and other differentially expressed genes (DEGs) in BRCA, PRAD, OV, and TGCT cancers (Fig. 9A-D, Fig. S5 A-D), revealing several significant findings. Among the top 20 positively correlated genes in the heatmaps (Fig. 9A-D), GPSM3 was consistently identified across BRCA, PRAD, and TGCT (Fig. 9A, B, D). GPSM3 has been identified as a prognostic and immunological biomarker in several cancers. It is mainly expressed in myeloid cells, playing crucial roles in regulating migration and inflammatory responses of these cells [63, 64, 65]. In BRCA and PRAD, UCP2 expression was significantly associated with two critical immune-related genes: CD74 and LGAL9 (Fig. 9A, B). CD74 is known to function as a receptor for macrophage migration inhibitory factor (MIF) and plays a pivotal role in antigen processing and immune responses [66, 67]. LGAL9 (galectin-9), on the other hand, is involved in immune regulation by modulating T-cell activity and contributing to immune tolerance [68, 69]. These findings suggest that UCP2 plays significant roles in immune activation and regulation in both male and female cancers, acting through critical immune modulators like GPSM3, CD74, and LGAL9.
In BRCA, gene ontology (GO) enrichment showed that UCP2 was involved in regulating leucocyte chemotaxis. GO enrichment from the GSEA database indicated that high UCP2 expression participated in the adaptive immune response and immune response regulating cell surface receptor signaling. GSEA-KEGG indicated UCP2 participation in cytokine-cytokine receptor interaction (Fig. 9A). In PRAD, GO enrichment associated UCP2 with leucocyte cell-cell adhesion. GSEA-GO analysis revealed that UCP2 was associated with the adaptive immune response (Fig. 9B). In OV, GSEA-GO analysis associated high UCP2 expression with improved cell-killing pathways, and GSEA-KEGG analysis connected UCP2 to immune response pathways, such as those involved in Epstein-Barr virus infection and NOD-like receptor signaling (Fig. 9C). In TGCT, leucocyte cell-cell adhesion genes were among the UCP2 positively co-expressed genes in GO. GO database from GSEA enrichment showed that UCP2 participated in the adaptive immune response, antigen receptor-mediated signaling pathway, immune response activating cells surface receptor signaling, leucocyte-mediated immunity, and lymphocyte-mediated immunity (Fig. 9D).
In addition, the analysis of UCP2 gene expression across these cancers revealed significant roles in multiple biological pathways. In breast cancer, UCP2 was associated with the Apoptosis, cell cycle, EMT, RTK, and PI3KAKT pathways, suggesting its influence on cell death, growth, and migration. In ovarian cancer, UCP2 negatively correlated with the EMT pathway, possibly affecting metastasis. In prostate and testicular cancers, UCP2 played a crucial role in hormone signaling, cell growth, and survival, particularly in the Hormone ER, PI3KAKT, TSCmTOR, and DNA damage pathways, indicating its broad involvement in cancer progression and regulation (Fig.S5E, Supplementary file 2). These data suggest that UCP2 is implicated in immune regulation, influencing immune-related pathways with significant associations observed in both male and female cancers.
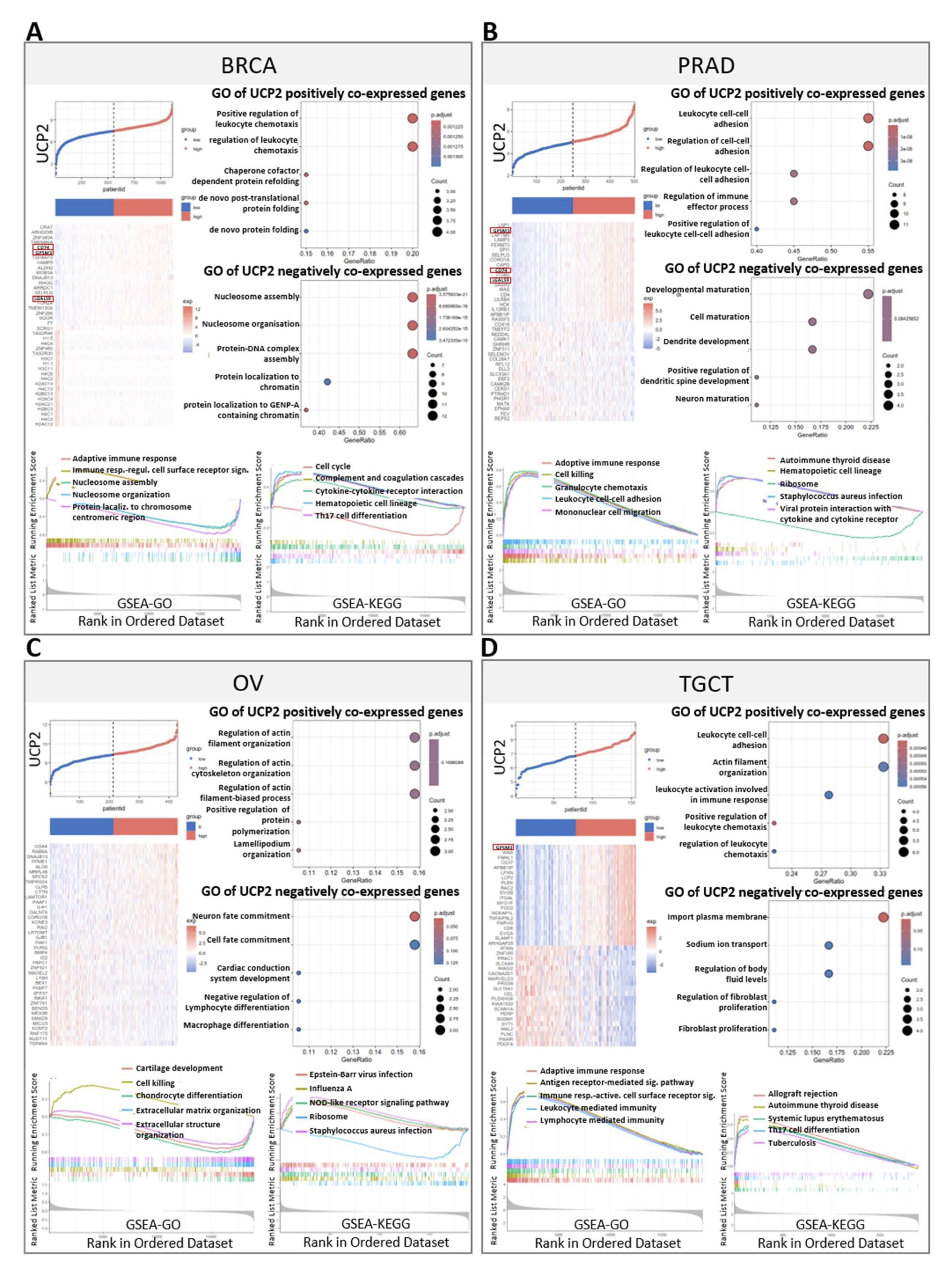
Fig. 9: DEGs analysis, GO analysis, GSEA-GO analysis, and GSEA-KEGG analysis in BRCA, PRAD, OV, and TGCT. Expression heatmap and enriched Gene Ontology (GO) pathways of genes significantly positively or negatively correlated with UCP2 in patients with BRCA (A), PRAD (B), OV (C), and TGCT (D). GSEA-GO and GSEA-KEGG analyses were conducted. Pearson correlation analysis was used to calculate the correlation between UCP2 expression and immune-related genes. Similar clusters were identified using the complete linkage method. Significance levels are indicated as **p < 0.01 and *p < 0.05. The heatmap was drawn using the TCGAplot R package (v7.0.1) (https://github.com/tjhwangxiong/TCGAplot).
UCP2 expression predicts drug sensitivity
Understanding the susceptibility of cancer cells to antineoplastic drugs is essential for developing effective anticancer therapies. The correlation between UCP2 and drug sensitivity was investigated in CTRP and GDSC databases using the Gene Set Cancer Analysis (Fig. 10A, B, Supplementary file 3). Drugs targeting critical oncogenic pathways and epigenetic regulators, including LRRK2, histone deacetylase, PI4KIIβ, and BET proteins, were negatively correlated with UCP2. On the other hand, UCP2 expression was positively correlated with seven specific drugs, such as 17-AAG, bleomycin, docetaxel, PD-0325901, RDEA119, selumetinib, and trametinib. These results indicate that UCP2 expression may predict treatment outcomes.
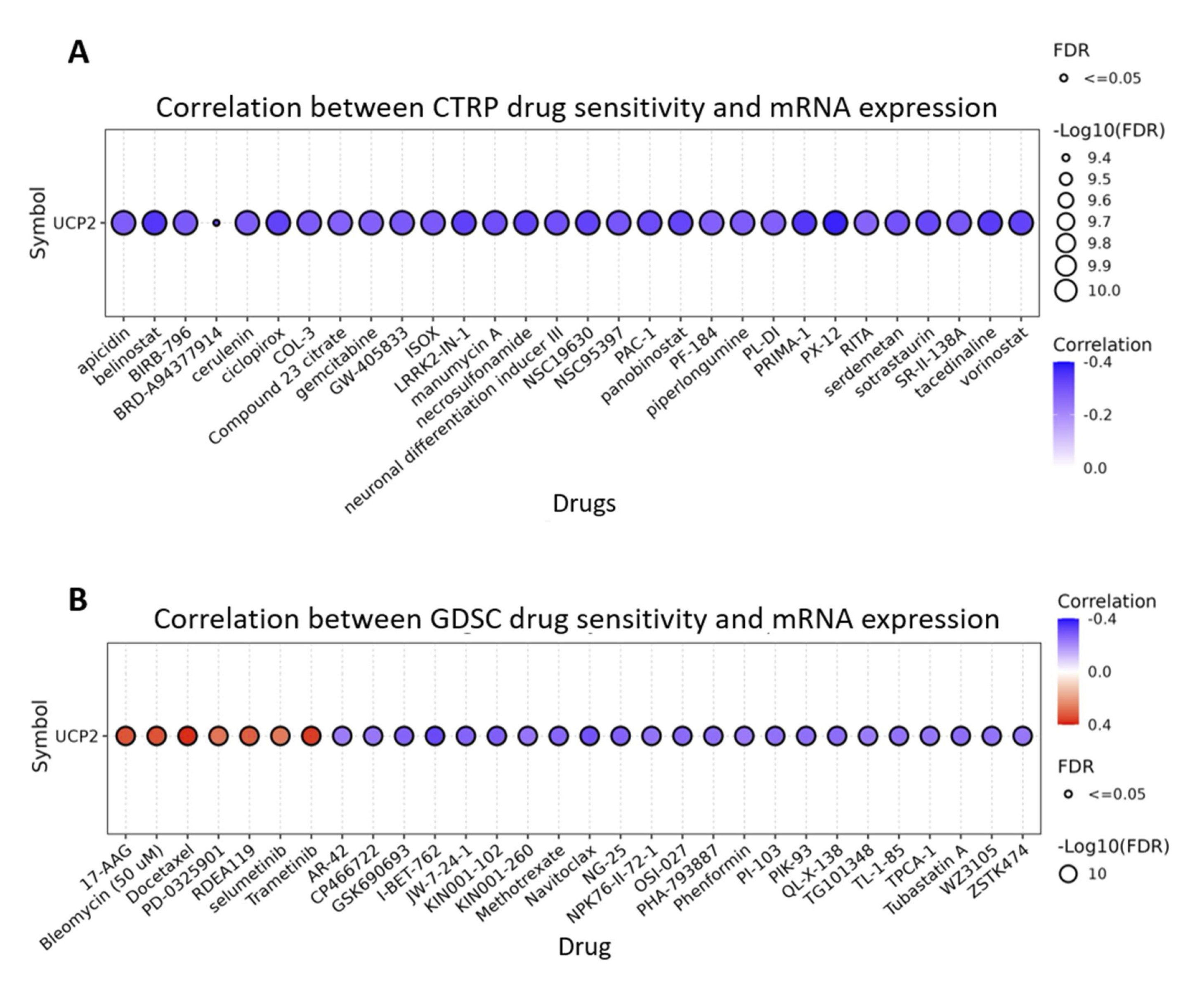
Fig. 10: UCP2 Expression Predicts Drug Sensitivity. (A) Correlation between UCP2 expression and the top 30 CTRP drug sensitivities in pan-cancer. (B) Correlation between UCP2 expression and the top 30 GDSC drug sensitivities in pan-cancer.
Discussion
Excessive ROS accumulation can trigger cancer by altering cellular metabolism and immune responses, enabling cancer cells to survive, multiply, and spread to other body parts. In this context, UCP2, known for its various functions in regulating oxidative stress and metabolism, emerges as a potential biomarker and therapeutic target in cancer. Depending on the type of tumor, its metabolic status, and its stage, UCP2 could be strategically targeted by either inhibiting or activating it to influence cancer outcomes [30]. In this study, we comprehensively examined the distinct characteristics of UCP2. This included analyzing its expression, genomic mutations, instability, prognosis, association with immune infiltration, and relationship with steroidogenic and sex receptor genes in human cancer, with a particular emphasis on hormone-driven cancers, such as breast (BRCA), prostate (PRAD), ovarian (OV), and testicular (TGCT) cancers.
UCP2 levels vary across cancer types, reflecting different functions and metabolic tumor characteristics [25],[26]. Our results showed that UCP2 was elevated in BRCA and OV, while reduced levels were observed in PRAD and TGCT, suggesting a potential sex-specific influence of UCP2 in these cancers. In addition to PRAD, reduced UCP2 expression was found in some cancers, with a higher incidence in males. These involve kidney chromophobe (KICH), lung squamous cell carcinoma (LUSC), and pancreatic adenocarcinoma (PAAD) (Table S5), [70, 71, 72]. Conversely, cancers such as BRCA, thyroid cancer (THCA), lung adenocarcinoma (LUAD), and uterine corpus endometrial carcinoma (UCEC) showed upregulated UCP2 levels and are more prevalent in females (Table S5). These results indicate that the expression of UCP2 plays a dual and context-dependent role in cancer, with potential gender-specific influence, highlighting the complexity of the UCP2 function and the need for further research. UCP2 plays a crucial role in regulating mitochondrial function, controlling ROS generation, and contributing to chemoresistance [73, 74, 17, 59]. UCP2 functions as a survival enhancer for various cancer cells, contributing to treatment resistance, and resulting in poor patient outcomes [60],[75], [76]. However, it may also, in specific contexts, be associated with improved prognosis [77], suggesting its involvement in distinct molecular mechanisms. The prognosis analysis presented in this study further underscores the dual role of UCP2 in hormone-related cancers such as breast, prostate, ovarian, and testicular cancers. UCP2 expression was associated with prolonged survival in ovarian cancer while contributing to reduced lifespan in prostate cancer.
The differences in UCP2 expression based on gender and their connection to survival indicate that sex hormones or related pathways may play a role in regulating UCP2 activity in cancer. For example, UCP2 has been identified as a risk factor in cancers with reduced UCP2 levels (Table S2), such as kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), and lower-grade glioma (LGG), which are more common in males (Table S5) [70, 78, 79]. Conversely, UCP2 predicted a better prognosis in cervical squamous cell carcinoma (CESC), lung adenocarcinoma (LUAD) [80], and OV. These malignancies exhibit upregulated UCP2 expression (Table S2) and are exclusively or more frequent in females (Table S5).
UCP2 gene amplification has been found in several hormone-driven cancers, with its polymorphism associated with increased disease risk [21],[22]. Our analysis showed dominant UCP2 amplifications in OV and BRCA and deletions in TGCT, indicating that UCP2 may play a distinct role in the oncogenic processes of these cancers and emphasize its potential involvement in gender-related tumor progression.
The connection between UCP2 and key immunotherapy biomarkers, such as tumor mutational burden (TMB) and microsatellite instability (MSI) [81], demonstrates its potential influence on tumor immunogenicity. Specifically, UCP2 showed a positive correlation with TMB in OV and a negative association with MSI in TGCT, suggesting that it could be a predictive biomarker for immunotherapy effectiveness in these malignancies.
UCP2 has been implicated in shaping the tumor microenvironment (TME), particularly in breast and melanoma cancers, through the modulation of immune cell infiltration and therapy resistance [60, 82]. Our data from breast, prostate, ovarian, and testicular cancer analysis revealed that UCP2 correlated with immune cell populations involved in immune evasion. The copy number variations (CNVs) of UCP2 significantly affected immune cell composition, particularly macrophages, T cells, and regulatory T cells in these cancers. Amplifications of UCP2 were correlated with increased macrophage infiltration in breast and prostate cancers. In breast cancer, UCP2 amplifications also increased B cell and dendritic cell infiltration, suggesting an anti-tumor inflammatory environment. Meanwhile, in prostate cancer, increased iTregs indicated a pro-tumor inflammatory environment.
In contrast, the deletions of UCP2 were linked to increased cytotoxic T cells in prostate and testicular cancers, promoting anti-tumor immunity. The consistent association between UCP2 expression and immune response pathways, including adaptive immunity, immune receptor signaling, leukocyte adhesion, immune cell migration, and immune activation, further supports the role of UCP2 in coordinating immune-related processes and influencing the TME.
Gender-related cancers are highly influenced by sex hormones, and disruption in steroidogenic pathways can lead to the development and progression of tumors [83]. CYP11A1, CYP19A1, and HSD17B2 are crucial enzyme genes in steroidogenesis, playing important roles in the biosynthesis of sex hormones such as estrogen and testosterone [84, 85, 86]. Changes in these enzymes are frequently observed in sex-dependent cancers, contributing to an imbalance in hormone production and regulation that drives tumor progression. UCP2 displayed strong positive correlations with these genes in breast and prostate cancer, potentially affecting cancer development and progression through its interaction with steroid hormone synthesis pathways. In addition, the correlation of UCP2 with estrogen and progesterone receptor genes in breast, ovarian, prostate, and testicular cancers, indicates a broader influence on hormone receptor signaling. Interestingly, the absence of a significant correlation between UCP2 and androgen receptor genes in male cancers suggests that UCP2 may have a more prominent role in estrogen than androgen receptor pathways. These findings highlight the context-dependent influence of UCP2 on steroid metabolism and hormone receptor activity, with potential implications for targeted therapeutic strategies.
Moreover, our analysis demonstrated that UCP2 expression predicts drug sensitivity, with high UCP2 levels correlating with resistance to several oncogenic therapies but increased sensitivity to agents such as 17-AAG, docetaxel, bleomycin, selumetinib, and trametinib. This suggests these drugs may be particularly effective in tumors with elevated UCP2 levels and combining them with UCP2 inhibitors could enhance cancer treatment. However, specific UCP2 inhibitors are not yet available for clinical use. Additionally, UCP2 involvement in steroidogenesis and hormone receptor signaling, particularly in hormone-dependent cancer, highlights the need to consider other combination therapies. In these cases, combining hormone-targeting treatments with UCP2-sensitive drugs may offer a more effective approach. For example, in breast and ovarian cancers, combining endocrine therapies (e.g., tamoxifen, aromatase inhibitors) with UCP2-sensitive drugs (e.g., docetaxel or 17-AAG) can target hormone receptor signaling and enhance cancer cell death [87, 88, 89, 90, 91]. In resistant cases, selective receptor degraders (SERDs; e.g., fulvestrant) combined with MEK inhibitors (e.g., trametinib or selumetinib) could address the UCP2-driven resistance mechanism [92][93]. In prostate and testicular cancers, UCP2 did not correlate with androgen receptors, so alternative pathways should be targeted. Combining CYP17A1 inhibitors (e.g., abiraterone) with MEK inhibitors could target both steroidogenesis and MAPK signaling [94]. Alternatively, estrogen receptor antagonists paired with UCP2-sensitive drugs [95, 96] may further improve outcomes in hormone receptor-positive, UCP2-high tumors. In addition to hormonal pathways, our findings indicate a link between UCP2 expression and immune checkpoint genes, suggesting another potential approach for combined therapy. Combining immunotherapies, such as PD-L1 or CTLA-4 inhibitors, hormone therapies (estrogen or androgen blockade), and UCP2-sensitive drugs, such as 17-AAG, could be a way to improve treatment strategies for tumors with high UCP2 expression.
Strengths and Limitations of Data Sources
This study used large-scale databases such as TCGA, cBioPortal, and GSCA to investigate UCP2 expression and its implications in cancer. These databases provide extensive genomic, transcriptomic, and clinical datasets, allowing for comprehensive multi-omics analyses across various types of cancer. Such an integrative approach is essential for identifying new biomarkers and therapeutic targets, such as UCP2, contributing to advances in precision oncology. TCGA has played a crucial role in discovering key oncogenes and tumor suppressors, which has, in turn, facilitated the development of targeted cancer therapies. Similarly, the GSCA has provided valuable insights into the relationship between gene expression, such as UCP2, and drug sensitivity, advancing more tailored and individualized treatment strategies. However, it is essential to consider several limitations. The lack of important clinical information, such as survival outcomes, cancer staging, and treatment regimens, limited the analysis. Moreover, genetic variations in non-coding, crucial for gene regulation and disease progression, were underrepresented. The lack of data on non-coding RNA, which plays a pivotal role in cancer-related gene regulation, further limited the ability to fully explore regulatory mechanisms such as those involving UCP2. Additionally, the lack of healthy control data and precise drug dosage information complicated the ability to conduct rigorous comparative analyses. Future research should prioritize integrating more comprehensive datasets, including clinical, regulatory, and pharmacological data, to enhance understanding of the oncogenic role of UCP2 and to optimize therapeutic approaches accordingly.
Conclusion
In conclusion, we conducted a comprehensive pan-cancer analysis of UCP2. The study revealed significantly increased UCP2 expression in most TCGA cancers, including breast, while observing markedly decreased expression in certain cancers, such as prostate cancer. We have demonstrated a potential correlation between UCP2 and prognosis in certain cancers. Additionally, we have identified a possible link between the prognostic significance of UCP2 expression and TMB, MSI, immune infiltration, and genes associated with immunotherapy. UCP2, indeed, is highly expressed in immune cells. Therefore, it is worth noting that there is a limitation in the dataset analysis, as it does not allow us to distinguish whether UCP2 expression originates from tumor cells or immune cells. Thus, additional research is required to clarify whether the observed UCP2 expression influences prognosis through its impact on cancer cell behavior or the increased presence of UCP2-expressing immune infiltrates. Finally, the involvement of UCP2 in shaping the tumor microenvironment, including its positive correlation with macrophage infiltration and immune-related pathways, highlights its potential to promote anti-tumor immune responses. Conversely, its reduced expression in cancers like the prostate and its association with worse prognosis in some contexts indicate its complex role in tumor biology, potentially linked to sex-specific mechanisms and hormonal pathways. Estrogens and androgens can regulate mitochondrial biogenesis, oxidative stress responses, and metabolic functions, all of which are tightly linked to UCP2 activity. These hormones might influence how UCP2 is amplified or deleted and its functional impact in different cancers. Understanding how sex hormones could modulate UCP2 expression and its genetic alterations and whether to which extent UCP2 affects sex hormone levels could provide insights into sex-specific cancer therapies and the distinct metabolic vulnerabilities of cancers in men versus women.
Acknowledgements
We thank I. Szabo for helpful discussions and critical reading of the manuscript.
Author Contributions
Conceptualization, S.S., V.C., and T.V.; methodology, software, validation, formal analysis, S.S.; data curation S.S., V.C., and T.V.; writing- original draft preparation S.S., V.C., and T.V.; supervision V.C. and T.V.; funding acquisition, V.C. and T.V.
Funding
T.V is grateful to the National Center for Gene Therapy and Drugs Based on RNA Technology, funded in the framework of the National Recovery and Resilience Plan (PNRR-CN3), Mission 4, Component 2, Investment 1.4, funded by the European Union—Next Generation EU, Project CN00000041, CUP C93C22002780006, CN3, Spoke n.2, Cancer. V.C. expresses gratitude for the financial support from the Project of National Interest Research 2022 (PRIN, grant number 2022ZY7ATN). S.S. expresses gratitude for the fellowship provided by Project of National Interest Research 2022 (PRIN, grant number 2022ZY7ATN). V.C. and T.V. are grateful for the financial support from DOR (Dotazione Ordinaria della Ricerca Dipartamentale) projects – Call 2023-2024.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
| 1 | Hanahan D. Hallmarks of Cancer: New Dimensions. Vol. 12, Cancer Discovery. 2022.
https://doi.org/10.1158/2159-8290.CD-21-1059 |
| 2 | Liberti M V., Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Vol. 41, Trends in Biochemical Sciences. 2016.
https://doi.org/10.1016/j.tibs.2016.01.004 |
| 3 | Liu Y, Sun Y, Guo Y, Shi X, Chen X, Feng W, Wu LL, Zhang J, Yu S, Wang Y, Shi Y. An Overview: The Diversified Role of Mitochondria in Cancer Metabolism. Vol. 19, International Journal of Biological Sciences. 2023.
https://doi.org/10.7150/ijbs.81609 |
| 4 | Roger AJ, Muñoz-Gómez SA, Kamikawa R. The Origin and Diversification of Mitochondria. Vol. 27, Current Biology. 2017.
https://doi.org/10.1016/j.cub.2017.09.015 |
| 5 | Bonora M, Patergnani S, Rimessi A, de Marchi E, Suski JM, Bononi A, Giorgi C, Marchi S, Missiroli S, Poletti F, Wieckowski MR, Pinton P. ATP synthesis and storage. Vol. 8, Purinergic Signalling. 2012.
https://doi.org/10.1007/s11302-012-9305-8 |
| 6 | Hong Y, Boiti A, Vallone D, Foulkes NS. Reactive Oxygen Species Signaling and Oxidative Stress: Transcriptional Regulation and Evolution. Vol. 13, Antioxidants. 2024.
https://doi.org/10.3390/antiox13030312 |
| 7 | Macherel D, Haraux F, Guillou H, Bourgeois O. The conundrum of hot mitochondria. Biochim Biophys Acta - Bioenerg. 2021;1862(2).
https://doi.org/10.1016/j.bbabio.2020.148348 |
| 8 | Cadenas S. Mitochondrial uncoupling, ROS generation and cardioprotection. Vol. 1859, Biochimica et Biophysica Acta - Bioenergetics. 2018.
https://doi.org/10.1016/j.bbabio.2018.05.019 |
| 9 | Chan CB, De Leo D, Joseph JW, McQuaid TS, Ha XF, Xu F, Tsushima RG, Pennefather PS, Salapatek AMF, Wheeler MB. Increased uncoupling protein-2 levels in β-cells are associated with impaired glucose-stimulated insulin secretion: Mechanism of action. Diabetes. 2001;50(6).
https://doi.org/10.2337/diabetes.50.6.1302 |
| 10 | Wang MY, Shimabukuro M, Lee Y, Trinh KY, Chen JL, Newgard CB, Unger RH. Adenovirus-mediated overexpression of uncoupling protein-2 in pancreatic islets of Zucker diabetic rats increases oxidative activity and improves β- cell function. Diabetes. 1999;48(5).
https://doi.org/10.2337/diabetes.48.5.1020 |
| 11 | Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, β cell dysfunction, and type 2 diabetes. Cell. 2001;105(6).
https://doi.org/10.1016/S0092-8674(01)00378-6 |
| 12 | Hong Y, Fink BD, Dillon JS, Sivitz WI. Effects of adenoviral overexpression of uncoupling protein-2 and -3 on mitochondrial respiration in insulinoma cells. Endocrinology. 2001;142(1):249-56.
https://doi.org/10.1210/endo.142.1.7889 |
| 13 | Nègre‐Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, Salvayre R, Pénicaud L, Casteilla L. A role for uncoupling protein‐2 as a regulator of mitochondrial hydrogen peroxide generation. FASEB J. 1997;11(10).
https://doi.org/10.1096/fasebj.11.10.9271366 |
| 14 | Pierelli G, Stanzione R, Forte M, Migliarino S, Perelli M, Volpe M, Rubattu S. Uncoupling protein 2: A key player and a potential therapeutic target in vascular diseases. Vol. 2017, Oxidative Medicine and Cellular Longevity. 2017.
https://doi.org/10.1155/2017/7348372 |
| 15 | Hass DT, Barnstable CJ. Uncoupling proteins in the mitochondrial defense against oxidative stress. Vol. 83, Progress in Retinal and Eye Research. 2021.
https://doi.org/10.1016/j.preteyeres.2021.100941 |
| 16 | Basu Ball W, Kar S, Mukherjee M, Chande AG, Mukhopadhyaya R, Das PK. Uncoupling Protein 2 Negatively Regulates Mitochondrial Reactive Oxygen Species Generation and Induces Phosphatase-Mediated Anti-Inflammatory Response in Experimental Visceral Leishmaniasis. J Immunol. 2011;187(3).
https://doi.org/10.4049/jimmunol.1004237 |
| 17 | Yu J, Shi L, Lin W, Lu B, Zhao Y. UCP2 promotes proliferation and chemoresistance through regulating the NF-κB/β-catenin axis and mitochondrial ROS in gallbladder cancer. Biochem Pharmacol. 2020;172.
https://doi.org/10.1016/j.bcp.2019.113745 |
| 18 | Kukat A, Dogan SA, Edgar D, Mourier A, Jacoby C, Maiti P, Mauer J, Becker C, Senft K, Wibom R, Kudin AP, Hultenby K, Flögel U, Rosenkranz S, Ricquier D, Kunz WS, Trifunovic A. Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity. PLoS Genet. 2014;10(6).
https://doi.org/10.1371/journal.pgen.1004385 |
| 19 | Bouillaud F. UCP2, not a physiologically relevant uncoupler but a glucose sparing switch impacting ROS production and glucose sensing. Vol. 1787, Biochimica et Biophysica Acta - Bioenergetics. 2009.
https://doi.org/10.1016/j.bbabio.2009.01.003 |
| 20 | Toda C, Diano S. Mitochondrial UCP2 in the central regulation of metabolism. Vol. 28, Best Practice and Research: Clinical Endocrinology and Metabolism. 2014.
https://doi.org/10.1016/j.beem.2014.02.006 |
| 21 | Donadelli M, Dando I, Fiorini C, Palmieri M. UCP2, a mitochondrial protein regulated at multiple levels. Vol. 71, Cellular and Molecular Life Sciences. 2014.
https://doi.org/10.1007/s00018-013-1407-0 |
| 22 | Li J, Jiang R, Cong X, Zhao Y. UCP2 gene polymorphisms in obesity and diabetes, and the role of UCP2 in cancer. Vol. 593, FEBS Letters. 2019.
https://doi.org/10.1002/1873-3468.13546 |
| 23 | Li W, Nichols K, Nathan CA, Zhao Y. Mitochondrial uncoupling protein 2 is up-regulated in human head and neck, skin, pancreatic, and prostate tumors. Cancer Biomarkers. 2013;13(5).
https://doi.org/10.3233/CBM-130369 |
| 24 | Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A, Thangaraj K, Buchsbaum DJ, LoBuglio AF, Singh KK. Cellular model of Warburg Effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS One. 2011;6(9).
https://doi.org/10.1371/journal.pone.0024792 |
| 25 | Esteves P, Pecqueur C, Ransy C, Esnous C, Lenoir V, Bouillaud F, Bulteau AL, Lombès A, Prip-Buus C, Ricquier D, Alves-Guerra MC. Mitochondrial retrograde signaling mediated by UCP2 inhibits cancer cell proliferation and tumorigenesis. Cancer Res. 2014;74(14).
https://doi.org/10.1158/0008-5472.CAN-13-3383 |
| 26 | Esteves P, Pecqueur C, Alves-Guerra MC. UCP2 induces metabolic reprogramming to inhibit proliferation of cancer cells. Mol Cell Oncol. 2015;2(1).
https://doi.org/10.4161/23723556.2014.975024 |
| 27 | Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, Paradies E, Scarcia P, Palmieri F, Bouillaud F, Fiermonte G. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci U S A. 2014;111(3).
https://doi.org/10.1073/pnas.1317400111 |
| 28 | Raho S, Capobianco L, Malivindi R, Vozza A, Piazzolla C, De Leonardis F, Gorgoglione R, Scarcia P, Pezzuto F, Agrimi G, Barile SN, Pisano I, Reshkin SJ, Greco MR, Cardone RA, Rago V, Li Y, Marobbio CMT, Sommergruber W, Riley CL, Lasorsa FM, Mills E, Vegliante MC, De Benedetto GE, Fratantonio D, Palmieri L, Dolce V, Fiermonte G. KRAS-regulated glutamine metabolism requires UCP2-mediated aspartate transport to support pancreatic cancer growth. Nat Metab. 2020;2(12).
https://doi.org/10.1038/s42255-020-00315-1 |
| 29 | Sancerni T, Renoult O, Luby A, Caradeuc C, Lenoir V, Croyal M, Ransy C, Aguilar E, Postic C, Bertho G, Dentin R, Prip-Buus C, Pecqueur C, Alves-Guerra MC. UCP2 silencing restrains leukemia cell proliferation through glutamine metabolic remodeling. Front Immunol. 2022;13.
https://doi.org/10.3389/fimmu.2022.960226 |
| 30 | Luby A, Alves-Guerra MC. UCP2 as a Cancer Target through Energy Metabolism and Oxidative Stress Control. Vol. 23, International Journal of Molecular Sciences. 2022.
https://doi.org/10.3390/ijms232315077 |
| 31 | Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Vol. 21, Cancer Cell. 2012.
https://doi.org/10.1016/j.ccr.2012.02.022 |
| 32 | Craig SG, Humphries MP, Alderdice M, Bingham V, Richman SD, Loughrey MB, Coleman HG, Viratham-Pulsawatdi A, McCombe K, Murray GI, Blake A, Domingo E, Robineau J, Brown L, Fisher D, Seymour MT, Quirke P, Bankhead P, McQuaid S, Lawler M, McArt DG, Maughan TS, James JA, Salto-Tellez M. Immune status is prognostic for poor survival in colorectal cancer patients and is associated with tumour hypoxia. Br J Cancer. 2020;123(8).
https://doi.org/10.1038/s41416-020-0985-5 |
| 33 | Botticelli A, Pomati G, Cirillo A, Scagnoli S, Pisegna S, Chiavassa A, Rossi E, Schinzari G, Tortora G, Di Pietro FR, Cerbelli B, Di Filippo A, Amirhassankhani S, Scala A, Zizzari IG, Cortesi E, Tomao S, Nuti M, Mezi S, Marchetti P. The role of immune profile in predicting outcomes in cancer patients treated with immunotherapy. Front Immunol. 2022;13.
https://doi.org/10.3389/fimmu.2022.974087 |
| 34 | Baba Y, Nakagawa S, Toihata T, Harada K, Iwatsuki M, Hayashi H, Miyamoto Y, Yoshida N, Baba H. Pan-immune-inflammation Value and Prognosis in Patients With Esophageal Cancer. Ann Surg Open. 2022;3(1).
https://doi.org/10.1097/AS9.0000000000000113 |
| 35 | Cheng WC, Tsui YC, Ragusa S, Koelzer VH, Mina M, Franco F, Läubli H, Tschumi B, Speiser D, Romero P, Zippelius A, Petrova T V., Mertz K, Ciriello G, Ho PC. Uncoupling protein 2 reprograms the tumor microenvironment to support the anti-tumor immune cycle. Nat Immunol. 2019;20(2).
https://doi.org/10.1038/s41590-018-0290-0 |
| 36 | van Dierendonck XAMH, Sancerni T, Alves-Guerra MC, Stienstra R. The role of uncoupling protein 2 in macrophages and its impact on obesity-induced adipose tissue inflammation and insulin resistance. J Biol Chem. 2020;295(51).
https://doi.org/10.1074/jbc.RA120.014868 |
| 37 | Monnier M, Paolini L, Vinatier E, Mantovani A, Delneste Y, Jeannin P. Antitumor strategies targeting macrophages: the importance of considering the differences in differentiation/polarization processes between human and mouse macrophages. Vol. 10, Journal for ImmunoTherapy of Cancer. 2022.
https://doi.org/10.1136/jitc-2022-005560 |
| 38 | Shi R, Tang YQ, Miao H. Metabolism in tumor microenvironment: Implications for cancer immunotherapy. Vol. 1, MedComm. 2020.
https://doi.org/10.1002/mco2.6 |
| 39 | Lang L, Zheng D, Jiang Q, Meng T, Ma X, Yang Y. Uncoupling protein 2 modulates polarization and metabolism of human primary macrophages via glycolysis and the NF‑κB pathway. Exp Ther Med. 2023;26(6).
https://doi.org/10.3892/etm.2023.12282 |
| 40 | Xu Y, Wang L, Cao S, Hu R, Liu R, Hua K, Guo Z, Di HJ, Hu Z. Genipin improves reproductive health problems caused by circadian disruption in male mice. Reprod Biol Endocrinol. 2020;18(1).
https://doi.org/10.1186/s12958-020-00679-9 |
| 41 | Pedersen SB, Bruun JM, Kristensen K, Richelsen B. Regulation of UCP1, UCP2, and UCP3 mRNA expression in brown adipose tissue, white adipose tissue, and skeletal muscle in rats by estrogen. Biochem Biophys Res Commun. 2001 Oct 19;288(1):191-7.
https://doi.org/10.1006/bbrc.2001.5763 |
| 42 | Sun Y, Bu LG, Wang B, Ren J, Li TY, Kong LL, Ni H. Expression and hormone regulation of UCP2 in goat uterus. Anim Reprod Sci. 2022;243.
https://doi.org/10.1016/j.anireprosci.2022.107015 |
| 43 | Liao C, Wang X. TCGAplot: an R package for integrative pan-cancer analysis and visualization of TCGA multi-omics data. BMC Bioinformatics. 2023;24(1).
https://doi.org/10.1186/s12859-023-05615-3 |
| 44 | Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20).
https://doi.org/10.1093/bioinformatics/btz210 |
| 45 | Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11).
https://doi.org/10.1158/1535-7163.MCT-17-0386 |
| 46 | Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Vol. 22, Clinical Cancer Research. 2016.
https://doi.org/10.1158/1078-0432.CCR-15-1678 |
| 47 | Tang T, Huang X, Zhang G, Hong Z, Bai X, Liang T. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Vol. 6, Signal Transduction and Targeted Therapy. 2021.
https://doi.org/10.1038/s41392-020-00449-4 |
| 48 | Chaudhuri L, Srivastava RK, Kos F, Shrikant PA. Uncoupling protein 2 regulates metabolic reprogramming and fate of antigen-stimulated CD8+ T cells. Vol. 65, Cancer Immunology, Immunotherapy. 2016.
https://doi.org/10.1007/s00262-016-1851-4 |
| 49 | Kim C, Jeong E, Lee Y Bin, Kim D. Steroidogenic cytochrome P450 enzymes as drug target. Vol. 40, Toxicological Research. Springer; 2024 p. 325-33.
https://doi.org/10.1007/s43188-024-00237-0 |
| 50 | Manna PR, Ahmed AU, Yang S, Narasimhan M, Cohen-Tannoudji J, Slominski AT, Pruitt K. Genomic profiling of the steroidogenic acute regulatory protein in breast cancer: In silico assessments and a mechanistic perspective. Cancers (Basel). 2019;11(5).
https://doi.org/10.3390/cancers11050623 |
| 51 | Blanco LZ, Kuhn E, Morrison JC, Bahadirli-Talbott A, Smith-Sehdev A, Kurman RJ. Steroid hormone synthesis by the ovarian stroma surrounding epithelial ovarian tumors: A potential mechanism in ovarian tumorigenesis. Mod Pathol. 2017;30(4).
https://doi.org/10.1038/modpathol.2016.219 |
| 52 | Poutanen M, Hagberg Thulin M, Härkönen P. Targeting sex steroid biosynthesis for breast and prostate cancer therapy. Vol. 23, Nature Reviews Cancer. 2023.
https://doi.org/10.1038/s41568-023-00609-y |
| 53 | Giovannelli P, Ramaraj P, Williams C. Editorial: Role of Sex Steroids and Their Receptor in Cancers. Vol. 13, Frontiers in Endocrinology. 2022.
https://doi.org/10.3389/fendo.2022.883229 |
| 54 | Liu Y, Jiang H, Xing FQ, Huang WJ, Mao LH, He LY. Uncoupling protein 2 expression affects androgen synthesis in polycystic ovary syndrome. Endocrine. 2013;43(3).
https://doi.org/10.1007/s12020-012-9802-0 |
| 55 | Ge H, Zhang F, Shan D, Chen H, Wang X, Ling C, Xi H, Huang J, Zhu C, Lv J. Effects of mitochondrial uncoupling protein 2 inhibition by genipin in human cumulus cells. Biomed Res Int. 2015;2015.
https://doi.org/10.1155/2015/323246 |
| 56 | Goldstone J V., Sundaramoorthy M, Zhao B, Waterman MR, Stegeman JJ, Lamb DC. Genetic and structural analyses of cytochrome P450 hydroxylases in sex hormone biosynthesis: Sequential origin and subsequent coevolution. Mol Phylogenet Evol. 2016;94.
https://doi.org/10.1016/j.ympev.2015.09.012 |
| 57 | Pillerová M, Borbélyová V, Hodosy J, Riljak V, Renczés E, Frick KM, Tóthová Ľ. On the role of sex steroids in biological functions by classical and non-classical pathways. An update. Vol. 62, Frontiers in Neuroendocrinology. 2021.
https://doi.org/10.1016/j.yfrne.2021.100926 |
| 58 | Teh BT, Fearon ER. Genetic and Epigenetic Alterations in Cancer. Abeloff's Clin Oncol. 2020 Jan 1;209-224.e2.
https://doi.org/10.1016/B978-0-323-47674-4.00014-1 |
| 59 | Pons DG, Nadal-Serrano M, Torrens-Mas M, Valle A, Oliver J, Roca P. UCP2 inhibition sensitizes breast cancer cells to therapeutic agents by increasing oxidative stress. Free Radic Biol Med. 2015;86.
https://doi.org/10.1016/j.freeradbiomed.2015.04.032 |
| 60 | Yu X, Shi M, Wu Q, Wei W, Sun S, Zhu S. Identification of UCP1 and UCP2 as Potential Prognostic Markers in Breast Cancer: A Study Based on Immunohistochemical Analysis and Bioinformatics. Front Cell Dev Biol. 2022;10.
https://doi.org/10.3389/fcell.2022.891731 |
| 61 | Won KY, Kim GY, Kim YW, Lim SJ, Song JY. Uncoupling protein 2 (UCP2) and p53 expression in invasive ductal carcinoma of breast. Korean J Pathol. 2010;44(6).
https://doi.org/10.4132/KoreanJPathol.2010.44.6.565 |
| 62 | Lakshminarasimhan R, Liang G. The role of DNA methylation in cancer. Adv Exp Med Biol. 2016;945.
https://doi.org/10.1007/978-3-319-43624-1_7 |
| 63 | Giguère PM, Billard MJ, Laroche G, Buckley BK, Timoshchenko RG, McGinnis MW, Esserman D, Foreman O, Liu P, Siderovski DP, Tarrant TK. G-protein signaling modulator-3, a gene linked to autoimmune diseases, regulates monocyte function and its deficiency protects from inflammatory arthritis. Mol Immunol. 2013;54(2).
https://doi.org/10.1016/j.molimm.2012.12.001 |
| 64 | Dang HH, Ta HDK, Nguyen TTT, Anuraga G, Wang CY, Lee KH, Le NQK. Identifying GPSM family members as potential biomarkers in breast cancer: A comprehensive bioinformatics analysis. Biomedicines. 2021;9(9).
https://doi.org/10.3390/biomedicines9091144 |
| 65 | Wang M, Jia J, Cui Y, Peng Y, Jiang Y. Molecular and clinical characterization of a novel prognostic and immunologic biomarker gpsm3 in low-grade gliomas. Brain Sci. 2021;11(11).
https://doi.org/10.3390/brainsci11111529 |
| 66 | Hong WC, Lee DE, Kang HW, Kim MJ, Kim M, Kim JH, Fang S, Kim HJ, Park JS. CD74 Promotes a Pro-Inflammatory Tumor Microenvironment by Inducing S100A8 and S100A9 Secretion in Pancreatic Cancer. Int J Mol Sci. 2023;24(16).
https://doi.org/10.3390/ijms241612993 |
| 67 | Wang J, Li X, Xiao G, Desai J, Frentzas S, Wang ZM, Xia Y, Li B. CD74 is associated with inflamed tumor immune microenvironment and predicts responsiveness to PD-1/CTLA-4 bispecific antibody in patients with solid tumors. Cancer Immunol Immunother. 2024;73(2).
https://doi.org/10.1007/s00262-023-03604-2 |
| 68 | Yang R, Sun L, Li CF, Wang YH, Yao J, Li H, Yan M, Chang WC, Hsu JM, Cha JH, Hsu JL, Chou CW, Sun X, Deng Y, Chou CK, Yu D, Hung MC. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun. 2021;12(1).
https://doi.org/10.1038/s41467-021-21099-2 |
| 69 | Lv Y, Ma X, Ma Y, Du Y, Feng J. A new emerging target in cancer immunotherapy: Galectin-9 (LGALS9). Vol. 10, Genes and Diseases. 2023.
https://doi.org/10.1016/j.gendis.2022.05.020 |
| 70 | Peired AJ, Campi R, Angelotti ML, Antonelli G, Conte C, Lazzeri E, Becherucci F, Calistri L, Serni S, Romagnani P. Sex and gender differences in kidney cancer: Clinical and experimental evidence. Vol. 13, Cancers. 2021.
https://doi.org/10.3390/cancers13184588 |
| 71 | May L, Shows K, Nana-Sinkam P, Li H, Landry JW. Sex Differences in Lung Cancer. Vol. 15, Cancers. 2023.
https://doi.org/10.3390/cancers15123111 |
| 72 | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022 CA Cancer J Clin. 2022;72(1).
https://doi.org/10.3322/caac.21708 |
| 73 | Baffy G. Uncoupling protein-2 and cancer. Vol. 10, Mitochondrion. 2010.
https://doi.org/10.1016/j.mito.2009.12.143 |
| 74 | Derdak Z, Mark NM, Beldi G, Robson SC, Wands JR, Baffy G. The mitochondrial uncoupling protein-2 promotes chemoresistance in cancer cells. Cancer Res. 2008;68(8).
https://doi.org/10.1158/0008-5472.CAN-08-0053 |
| 75 | Yu J, Shi L, Shen X, Zhao Y. UCP2 regulates cholangiocarcinoma cell plasticity via mitochondria-to-AMPK signals. Biochem Pharmacol. 2019;166.
https://doi.org/10.1016/j.bcp.2019.05.017 |
| 76 | Song C, Liu Q, Qin J, Liu L, Zhou Z, Yang H. UCP2 promotes NSCLC proliferation and glycolysis via the mTOR/HIF-1α signaling. Cancer Med. 2024;13(3).
https://doi.org/10.1002/cam4.6938 |
| 77 | Aguilar E, Esteves P, Sancerni T, Lenoir V, Aparicio T, Bouillaud F, Dentin R, Prip-Buus C, Ricquier D, Pecqueur C, Guilmeau S, Alves-Guerra MC. UCP2 Deficiency Increases Colon Tumorigenesis by Promoting Lipid Synthesis and Depleting NADPH for Antioxidant Defenses. Cell Rep. 2019;28(9).
https://doi.org/10.1016/j.celrep.2019.07.097 |
| 78 | Stabellini N, Tomlinson B, Cullen J, Shanahan J, Waite K, Montero AJ, Barnholtz-Sloan JS, Hamerschlak N. Sex differences in adults with acute myeloid leukemia and the impact of sex on overall survival. Cancer Med. 2023;12(6).
https://doi.org/10.1002/cam4.5461 |
| 79 | Tewari S, Tom MC, Park DYJ, Wei W, Chao ST, Yu JS, Suh JH, Kilic S, Peereboom DM, Stevens GHJ, Lathia JD, Prayson R, Barnett GH, Angelov L, Mohammadi AM, Ahluwalia MS, Murphy ES. Sex-Specific Differences in Low-Grade Glioma Presentation and Outcome. Int J Radiat Oncol Biol Phys. 2022;114(2).
https://doi.org/10.1016/j.ijrobp.2022.05.036 |
| 80 | Wen H, Lin X, Sun D. Gender-specific nomogram models to predict the prognosis of male and female lung adenocarcinoma patients: a population-based analysis. Ann Transl Med. 2021;9(22).
https://doi.org/10.21037/atm-21-5367 |
| 81 | Palmeri M, Mehnert J, Silk AW, Jabbour SK, Ganesan S, Popli P, Riedlinger G, Stephenson R, de Meritens AB, Leiser A, Mayer T, Chan N, Spencer K, Girda E, Malhotra J, Chan T, Subbiah V, Groisberg R. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open. 2022;7(1).
https://doi.org/10.1016/j.esmoop.2021.100336 |
| 82 | Cheng WC, Tsui YC, Ragusa S, Koelzer VH, Mina M, Franco F, Läubli H, Tschumi B, Speiser D, Romero P, Zippelius A, Petrova T V., Mertz K, Ciriello G, Ho PC. Uncoupling protein 2 reprograms the tumor microenvironment to support the anti-tumor immune cycle. Nat Immunol. 2019 Feb 1;20(2):206-17.
https://doi.org/10.1038/s41590-018-0290-0 |
| 83 | Anderson AC, Acharya N. Steroid hormone regulation of immune responses in cancer. Vol. 4, Immunometabolism (United States). 2022.
https://doi.org/10.1097/IN9.0000000000000012 |
| 84 | Fan Z, Wang Z, Chen W, Cao Z, Li Y. Association between the CYP11 family and six cancer types. Oncol Lett. 2016;12(1).
https://doi.org/10.3892/ol.2016.4567 |
| 85 | Hong Y, Li H, Yuan YC, Chen S. Molecular characterization of aromatase. In: Annals of the New York Academy of Sciences. 2009.
https://doi.org/10.1111/j.1749-6632.2009.03703.x |
| 86 | Mindnich R, Möller G, Adamski J. The role of 17 beta-hydroxysteroid dehydrogenases. Vol. 218, Molecular and Cellular Endocrinology. 2004.
https://doi.org/10.1016/j.mce.2003.12.006 |
| 87 | Tremont A, Lu J, Cole JT. Endocrine therapy for early breast cancer: Updated review. Vol. 17, Ochsner Journal. 2017.
|
| 88 | Langdon SP, Gourley C, Gabra H, Stanley B. Endocrine therapy in epithelial ovarian cancer. Vol. 17, Expert Review of Anticancer Therapy. 2017.
https://doi.org/10.1080/14737140.2017.1272414 |
| 89 | Brown I, Shalli K, McDonald SL, Moir SE, Hutcheon AW, Heys SD, Schofield AC. Reduced expression of p27 is a novel mechanism of docetaxel resistance in breast cancer cells. Breast Cancer Res. 2004;6(5).
https://doi.org/10.1186/bcr918 |
| 90 | De Mattos-Arruda L, Cortes J. Breast cancer and HSP90 inhibitors: Is there a role beyond the HER2-positive subtype? Breast. 2012;21(4).
https://doi.org/10.1016/j.breast.2012.04.002 |
| 91 | Ikeda H, Taira N, Nogami T, Shien K, Okada M, Shien T, Doihara H, Miyoshi S. Combination treatment with fulvestrant and various cytotoxic agents (doxorubicin, paclitaxel, docetaxel, vinorelbine, and 5-fluorouracil) has a synergistic effect in estrogen receptor-positive breast cancer. Cancer Sci. 2011;102(11).
https://doi.org/10.1111/j.1349-7006.2011.02050.x |
| 92 | Bussies PL, Schlumbrecht M. Dual Fulvestrant-Trametinib Therapy in Recurrent Low-Grade Serous Ovarian Cancer. Oncologist. 2020;25(7).
https://doi.org/10.1634/theoncologist.2020-0101 |
| 93 | Zaman K, Winterhalder R, Mamot C, Hasler-Strub U, Rochlitz C, Mueller A, Berset C, Wiliders H, Perey L, Rudolf CB, Hawle H, Rondeau S, Neven P. Fulvestrant with or without selumetinib, a MEK 1/2 inhibitor, in breast cancer progressing after aromatase inhibitor therapy: A multicentre randomised placebo-controlled double-blind phase II trial, SAKK 21/08 Eur J Cancer. 2015;51(10).
https://doi.org/10.1016/j.ejca.2015.03.016 |
| 94 | Nickols NG, Nazarian R, Zhao SG, Tan V, Uzunangelov V, Xia Z, Baertsch R, Neeman E, Gao AC, Thomas G V., Howard L, De Hoedt AM, Stuart J, Goldstein T, Chi K, Gleave ME, Graff JN, Beer TM, Drake JM, Evans CP, Aggarwal R, Foye A, Feng FY, Small EJ, Aronson WJ, Freedland SJ, Witte ON, Huang J, Alumkal JJ, Reiter RE, Rettig MB. MEK-ERK signaling is a therapeutic target in metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis. 2019;22(4).
https://doi.org/10.1038/s41391-019-0134-5 |
| 95 | Gasent Blesa JM, Alberola Candel V. PSA decrease with fulvestrant acetate in a hormone-resistant metastatic prostate cancer patient. Onkologie. 2010;33(1-2).
https://doi.org/10.1159/000264612 |
| 96 | Murray V, Chen JK, Chung LH. The interaction of the metallo-glycopeptide anti-tumour drug bleomycin with DNA. Vol. 19, International Journal of Molecular Sciences. 2018.
https://doi.org/10.3390/ijms19051372 |